Adolescent alcohol exposure changes RNA modifications in adult brain by mass spectrometry-based comprehensive profiling analysis
2024-04-06YingYingChenZhuGuiDiHuMengYunChenJinGngHeSiYuYuYuQiFengJieWngBiFengYun
Ying-Ying Chen ,Zhu Gui ,Di Hu ,Meng-Yun Chen ,Jin-Gng He ,Si-Yu Yu ,Yu-Qi Feng,Jie Wng,f,g,∗,Bi-Feng Yun,,c,∗
a College of Chemistry and Molecular Sciences,Research Center of Public Health,Renmin Hospital of Wuhan University,Wuhan University,Wuhan 430072,China
b Department of Radiation and Medical Oncology,Zhongnan Hospital of Wuhan University,School of Public Health,Wuhan University,Wuhan 430071,China
c Cancer Precision Diagnosis and Treatment and Translational Medicine Hubei Engineering Research Center,Zhongnan Hospital of Wuhan University,Wuhan Research Center for Infectious Diseases and Cancer,Chinese Academy of Medical Sciences,Wuhan 430071,China
d Key Laboratory of Magnetic Resonance in Biological Systems,State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics,National Center for Magnetic Resonance in Wuhan,Wuhan Institute of Physics and Mathematics,Innovation Academy for Precision Measurement Science and Technology,Chinese Academy of Sciences-Wuhan National Laboratory for Optoelectronics,Wuhan 430071,China
e Phase I Clinical Trial Laboratory,Zhongnan Hospital of Wuhan University,Wuhan 430071,China
f Institute of Neuroscience and Brain Diseases,Xiangyang Central Hospital,Affiliated Hospital of Hubei University of Arts and Science,Xiangyang 441000,China
g University of Chinese Academy of Sciences,Beijing 101408,China
Keywords: Mass spectrometry RNA modification Adolescence Alcohol exposure Brain
ABSTRACT Alcohol consumption is one of the leading causes of death worldwide.Adolescence is a critical period of structural and functional maturation of the brain.Adolescent alcohol use can alter epigenetic modifications.However,little is known on the long-term effects of alcohol consumption during adolescence on RNA epigenetic modifications in brain.Herein,we systematically explored the long-term effects of alcohol exposure during adolescence on small RNA modifications in adult rat brain tissues by comprehensive liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis.We totally detected 26 modifications in small RNA of brain tissues.Notably,we observed most of these modifications were decreased in brain tissues.These results suggest that alcohol exposure during adolescence may impose a long-lasting impact on RNA modifications in brain tissues.This is the first report that alcohol use during adolescence can alter RNA modifications in adult brain.Collectively,this study suggests a long-term adverse effects of alcohol consumption on brain from RNA epigenetics angle by comprehensive mass spectrometry analysis.
Alcohol consumption is known to increase the risk of a number of human diseases [1],including mental disorders [2],liver diseases [3],cardiovascular diseases [4] and cancers [5].Alcohol use frequently emerge in adolescence,a crucial period for brain development.Increasing evidence has shown that alcohol use during adolescence can affect the structure and function of the brain[6].The consequences include a decrease in gray-matter volume[7],cortical gray-matter thickness [8],and disrupted white-matter integrity [9],along with an adverse influence on the development of attention,verbal learning,and memory [10].
Nucleic acid modifications are considered to play critical roles in diverse physiological processes [11–18].Alcohol exposure during adolescence can lead to dysregulation in DNA methylation[19].Like DNA modifications,RNA modifications have been recently viewed as new regulators in modulating gene expression [20].So far,more than 150 chemical modifications have been identified in diverse RNA species [21–24].RNA modifications are prevalent in small RNAs (<200 nt in length),including transfer RNAs (tRNAs),micro RNAs (miRNAs),and Piwi-interacting RNAs (piRNAs) [25–27].Many RNA modifications are dynamic and reversible,and play critical roles in the regulation of RNA generation,stability,folding,transportation,and metabolization [28,29].RNA modifications in tRNA will promote decoding ability and ensure translation accuracy [30].
Our group previously demonstrated that adolescent alcohol exposure could affect RNA modifications [31].However,this study only focuses on the short-term effect of alcohol exposure in RNA modifications in peripheral blood.Moreover,the long-term impacts of adolescent alcohol consumption on RNA modifications in brain are still lacked.
Here we explored the long-lasting effect of alcohol exposure during adolescence on the small RNA modifications in brain.In this respect,adolescent male Sprague Dawley (SD) rats (n=4/group)were intraperitoneally injected with ethanol (2 g/kg,20% w/v) or equivalent volume of saline during the post-natal day (PND) 34 to 46 with an interval of one day,and for a total of 7 times(Fig.1A).The alcohol exposure experiments were carried out according to precious reports with slight modification [19,31].At PND 77 (i.e.,the rats became adults),the rats were sacrificed and different brain tissues were collected to isolate the small RNA (<200 nt),which were further enzymatically digested followed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis (Fig.1B).The isolated small RNAs were examined by polyacrylamide gel electrophoresis (Fig.S1 in Supporting information).The animal experiment was approved by the animal care and use committee in the Innovation Academy for Precision Measurement Science and Technology,the Chinese Academy of Sciences (NO: APM21001A).
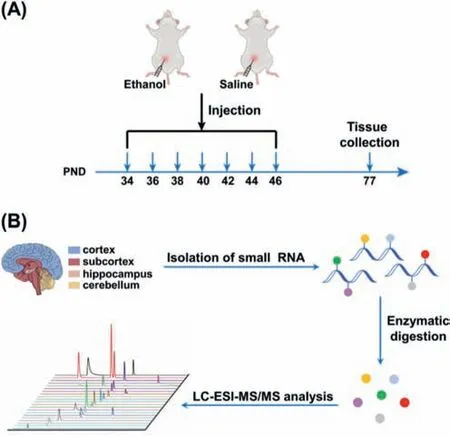
Fig.1.Schematic illustration for the alcohol exposure and mass spectrometry profiling of RNA modifications.(A) The schematic diagram of the timeline for the alcohol exposure and tissue collection from rats.(B) Workflow of profiling of small RNA modifications in four rat brain tissues of cortex,subcortex,hippocampus,and cerebellum by LC-ESI-MS/MS analysis.PND: postnatal day.
We first established the LC-ESI-MS/MS method for the simultaneous and comprehensive profiling of RNA modifications.After careful optimization of the chromatographic separation conditions,we could achieve the simultaneous determination of 51 nucleosides (structures are shown in Fig.S2 in Supporting information) with a single LC-ESI-MS/MS analysis.Specifically,eight isomeric RNA modification groups (N1-methyladenosine (m1A) andN6-methyladenosine (m6A);N1,N6-dimethyladenosine (m1,6A),N6,N6-dimethyladenosine (m6,6A)andN6,2′-O-dimethyladenosine (m6Am);5,2′-O-dimethylcytidine(m5Cm) and 3,2′-O-dimethylcytidine (m3Cm);1-methylguanosine(m1G),N2-methylguanosine (m2G) andN7-methylguanosine(m7G);3-methyluridine (m3U) and 5-methyluridine (m5U);cis-Zeatin-riboside (io6A (cis)) andtrans-Zeatin-riboside (io6A (trans));2-thiouridine (s2U) and 4-thiouridine (s4U);1-methylinosine(m1I) and 2′-O-methylinosine (Im)) and seven groups of RNA modifications with similar molecular weights (m1A,m6A and m1I;2′-O-methylguanosine (Am) and Im;2′-O-methylcytidine(Cm) and 2′-O-methyluridine (Um);m5Cm,m3Cm and 5,2′-Odimethyluridine (m5Um);m3U,m5U and 2-thiocytidine (s2C);s2C,5-hydroxyuridine (ho5U),s2U and s4U;5-hydroxymethylcytidine(hm5C),5-methoxyuridine (mo5U) and 5-methyl-2-thiouridine(m5s2U)) can be well distinguished (Table S2 in Supporting information).With the established LC-ESI-MS/MS method,we then analyzed modifications from small RNA of rat tissues.The results showed that 26 RNA modifications (Am,m6A,m1A,m1,6A,m6,6A,m6Am,N6-threonylcarbamoyladenosine (t6A),N6-isopentenyladenosine (i6A),inosine (I),m1I,Cm,m5Cm,m3Cm,5-formyl-2′-O-methylcytidine (f5Cm),N4-acetylcytidine (ac4C),2′-Omethylguanosine (Gm),m1G,m2G,m7G,N2,N2-dimethylguanosine(m2,2G),N2,N2,7-trimethylguanosine (m2,2,7G),Um,pseudouridine(Y),m5U,m5Um and 5-methoxycarbonylmethyluridine (mcm5U))could be detected from cortex,subcortex,hippocampus,and cerebellum (Fig.2 and Figs.S3-S14 in Supporting information).m3Cm and m1,6A,which were first reported by our group [32,33],were also detected in brain tissues.
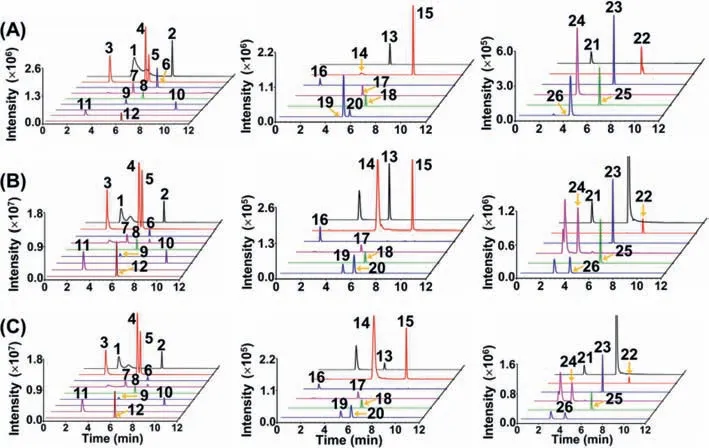
Fig.2.Extracted-ion chromatograms of detected 26 RNA modifications from rat cortex.(A) Nucleoside standards.(B) Nucleosides detected in small RNA of rat cortex from control group.(C) Nucleosides detected in small RNA of rat cortex from alcohol exposure group.The peaks were labeled with numbers.1,m1A;2,m6A;3,m7G;4,m1G;5,m2G;6,Am;7,m1I;8,m2,2,7G;9,Gm;10,i6A;11,Cm;12,m2,2G;13,ac4C;14,m1,6A;15,m6,6A;16,Y;17,mcm5U;18,f5Cm;19,m5Cm;20,m3Cm;21,I;22,m6Am;23,t6A;24,m5U;25,m5Um;26,Um.
To assess the abundance of modified nucleosides in small RNA of rat brain tissues,we generated calibration curves of these 26 modifications.In this regard,different amounts of nucleoside standards mixed with a given amount of isotopic internal standard (rC-13C5) were utilized to construct the calibration curves by plotting the peak area ratios (nucleosides/rC-13C5)versusthe amounts of nucleosides (Table S3 in Supporting information).The results indicated that good linearities were obtained and the coefficients of determination (R2) were higher than 0.99.The limits of detection(LODs) are defined as the amounts of the analytes at signal-tonoise ratio of 3.The LODs of the detected modifications ranged from 0.04 fmol to 213.37 fmol (Table S3).It can be seen that three uridine modifications (Um,Y and m5U) have higher LODs than other modifications,which is due to the low ionization efficiencies of uridine modifications in LC-ESI-MS/MS analysis.The relative errors (REs) and intra and inter-day relative standard deviations(RSDs) were calculated to evaluate the accuracy and precision of the method.The results showed the REs and RSDs were less than 14.3% and 12.7%,respectively (Table S4 in Supporting information),demonstrating good accuracy and precision of the method.Using the calibration curves,we successfully quantified these 26 modifications in small RNA of rat brain tissues,including cortex,subcortex,hippocampus,and cerebellum (Table S5 in Supporting information).The levels of these modifications are generally comparable among these four brain tissues (Fig.3).

Fig.3.The relative levels of small RNA modifications among rat brain tissues (cortex,subcortex,hippocampus,and cerebellum) from control group.
We next investigated the effect of adolescent alcohol exposure on small RNA modifications in brain tissues.The quantification results demonstrated an overall decreased trend of small RNA modifications in cortex,subcortex,hippocampus,and cerebellum(Fig.4 and Figs.S15-S17 in Supporting information).Specifically,fourteen,seven,five,and six modifications in rat cortex,subcortex,hippocampus,and cerebellum,respectively,were significantly decreased,and one modification in rat subcortex was significantly increased upon alcohol exposure (Fig.5).Notably,i6A was significantly decreased in all the four brain tissues;m6Am,ac4C and Am were significantly decreased in cortex,subcortex,and cerebellum;m7G and m2,2G were markedly decreased in cortex,hippocampus,and cerebellum.
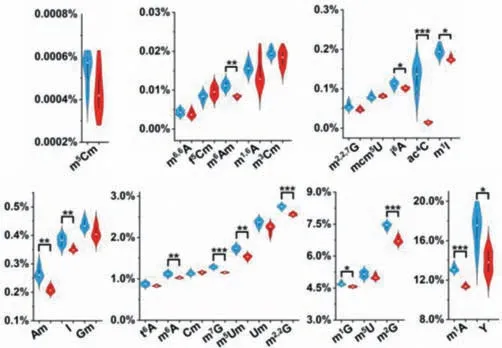
Fig.4.The measured levels of small RNA modifications in cortex from control and alcohol exposure groups.Shown in blue and red represent the modifications from control and alcohol exposure groups,respectively.The y-axis represents the levels of RNA modifications over their corresponding normal nucleosides.∗, P < 0.05;∗∗,P < 0.01;∗∗∗, P < 0.001.
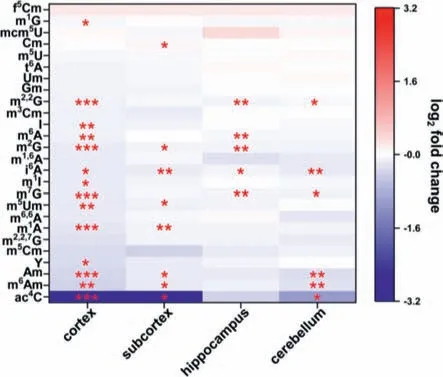
Fig.5.Long-term effect of alcohol use during adolescence on small RNA modifications in rat brain tissues.∗, P < 0.05;∗∗, P < 0.01;∗∗∗, P < 0.001.
Previous study showed that tRNA was the majority component(∼90%) in small RNA (<200 nt) [34].Thus,the detected modifications should be mainly from tRNA.i6A exists in the position 37 at the anticodon loops of a subset of cytosolic and mitochondrial tRNAs [35].i6A in tRNAs can promote translational efficiency and fidelity [35].It is worth noting that i6A was significantly decreased in all the four brain tissues upon alcohol exposure.We reason that the decreased level of i6A may lead to the attenuated translation and fidelity and eventually may cause neuronal diseases.m6A is the most important RNA modification and plays critical functions in a variety of physiological processes [36].Previous studies have shown that m6A is involved in neurodevelopment and memory deficits [37,38].It is therefore possible that the significantly decreased level of m6A in both cortex and hippocampus with alcohol use during adolescent could persist a long-term detrimental effect to memory.Adenosine-to-inosine (A-to-I) editing in RNA is prevalent in mammals [39–42],which is essential for maintaining normal neurodevelopment and function [43].The significantly decreased level of I in cortex may affect translation accuracy and eventually may affect the development of nervous system.Collectively,the overall declined trend of RNA modifications in brain documents that adolescent alcohol exposure alters small RNA modifications in adult brain.
In summary,we systematically investigated the long-term effect of adolescent alcohol exposure on small RNA modifications in brain by LC-ESI-MS/MS analysis.The quantification results show that adolescent alcohol exposure could change small RNA modifications in adult rat brain tissues,with an overall decline trend of RNA modifications in cortex,and subcortex,hippocampus and cerebellum.This study highlights that alcohol consumption during adolescence may impose a long-term effect on brain through reshaping the small RNA modifications,which should be valuable to elucidate the effects of alcohol-induced neurological diseases from the RNA epigenetics angle.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work is supported by the National Key R&D Program of China (Nos.2022YFA0806601,2022YFC3400700),the National Natural Science Foundation of China (Nos.22277093,22074110,21721005),the Interdisciplinary Innovative Talents Foundation from Renmin Hospital of Wuhan University (No.JCRCGW-2022–008),and the Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (No.ZNJC202208).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2023.108522.
杂志排行
Chinese Chemical Letters的其它文章
- Spin switching in corrole radical complex
- Benzothiadiazole-based materials for organic solar cells
- Mono-functionalized pillar[n]arenes: Syntheses,host–guest properties and applications✰
- Recent advances in two-step energy transfer light-harvesting systems driven by non-covalent self-assembly✩
- From oxygenated monomers to well-defined low-carbon polymers
- Doping-induced charge transfer in conductive polymers
