Single nucleotide polymorphism within chromosome 8q24 is associated with prostate cancer development in Saudi Arabia
2024-02-25AwdElsidOsmnShrAlhrbiAtifAliAhmedAsimAliElbgir
Awd Elsid Osmn ,Shr Alhrbi ,Atif Ali Ahmed,Asim Ali Elbgir
a Pathology and Clinical Laboratory Management Department(PCLM),King Fahad Medical City,Riyadh,Saudi Arabia
b Department of Pathology and Laboratory Medicine,University of Missouri at Children’s Mercy Hospital,Kansas City,MO,USA
KEYWORDS Prostate cancer;Saudi;Single nucleotide polymorphism;Allele
Abstract Objective: Genome-wide association studies have demonstrated that single nucleotide polymorphisms(SNPs)are important risk factors for the development of prostate cancer(PCa).Preliminary studies have suggested that the incidence of PCa in Saudi males is low but is probably familial or genetically related.Methods: To identify any possible association of SNP with PCa development in Saudi patients,we investigated a group of SNPs in Saudi PCa patients (n=85) and compared the outcomes to healthy normal controls(n=115)and nodular hyperplasia patients(n=120).DNA was extracted from paraffin-embedded formalin fixed tissue or whole blood from both patients’ groups and healthy control group.A total of thirteen SNPs were genotyped using TaqMan® minor groove binder polymerase chain reaction assay.Results: The rs16901979A,s629242T and rs1447295A alleles were found at significantly higher frequency in PCa patients than controls (p<0.05).The rs16901979 CA genotype was found at significantly greater frequency in PCa patients than in healthy controls (43% vs. 14%,odds ratio=4.6, p=0.0001) and benign hyperplasia group (43% vs. 25%,odds ratio=2.2, p=0.009).Conclusion: Our study has highlighted the association of rs16901979 SNP with PCa in Saudi males.Such findings have important implications in the PCa diagnosis and in screening unaffected family members of Saudi patients.
1.Introduction
Prostate cancer (PCa) is a predominant cancer disease among men worldwide and has the third highest mortality rate [1,2]with the mean age of 72 years for diagnosis [3].The International Agency for Research on Cancer and the World Cancer Research Fund and American Institute for Cancer Research have established a group of risk factors for PCa.Controlling tobacco products,changing dietary pattern,increasing physical activity,and controlling the obesity epidemic are the major opportunities in minimizing the global incidence and mortality of cancers [4,5].There are worldwide variations in PCa incidence rates.PCa in the United States is more frequent among African Americans than in Caucasians or Hispanics[6]and is also more common in males with a positive family history[7].Limited data exist on the prevalence of PCa in Saudi males,which is attributed to poor epidemiology estimation reports.However,the general clinical impression is that PCa prevalence is much lower in Saudi Arabia than in industrialized countries.The incidence rate in Saudi Arabia varies according to geographical regions but is estimated to be 6.1 per 100 000 individuals [8].A report by Almutairi et al.[9]in 2019 has shown the PCa prevalence among Saudi males is more than what is estimated by the Saudi Cancer Registry and is comparable to the prevalence in the developed countries.A regular screen for prostate-specific antigen among Saudi males is recommended [9].
Many studies carried out worldwide have suggested that PCa seems to run in families and may be inherited or genetic in origin.Germline mutations in the DNA damage repair genes (BRCA1,BRCA2,CHEK2,ATM,andPALB2) and DNA mismatch repair genes(MLH1,MSH2,MSH6,andPMS2)are risk factors that can drive the development of hereditary PCa types [10,11].Yadav et al.[12]conducted exome sequencing of 124 DNA damage repair and response genes in tumors from formalin-fixed,paraffin-embedded samples and compared them to normal tissue samples in African Americans and Caucasians.They identified 671 somatic mutations in tumors from African Americans and 762 somatic mutations in Caucasians.The predominant mutations includedEXO1,ATR,POLQ,NEIL3,ERCC6,BRCA2,BRCA1,XPC,JAG1,RPA1,POLE,ATM,andLIG1genes in African American men,andPOLQ,NEIL3,POLB,BRCA2,EXO1,ERCC6,ATR,RBBP8,BRCA1,ATM,JAG1,XPC,andPOLEgenes in Caucasians [12].
Several genome-wide association studies have confirmed the role of single nucleotide polymorphisms(SNPs)as factors associated with the development of PCa[13,14].Carriers of the homozygous T allele of the rs10993994 SNP were found at a higher risk for PCa than those having the homozygous C allele [15].Although few studies have reported on the epidemiology and genetics of PCa in the Middle East and Arab countries,the prevalence of SNP has not been previously adequately studied in Saudi Arabia.Accordingly,we have investigated a group of SNPs (Table 1) that were found in association with PCa in other populations to determine a possible association with PCa in Saudi patients.
2.Patients and methods
2.1.Patients and control
DNA was extracted from 320 individuals recruited to the study,including 85 patients with PCa,120 patients with benign hyperplasia(BH),and 115 healthy controls.Archived formalin-fixed paraffin embedded tissue was obtained from patients with PCa or nodular hyperplasia.On the other hand,peripheral blood in ethylene diamine tetra acetic acid was obtained from healthy unrelated blood donors with no history of PCa or other prostatic diseases.The study was approved by the Institutional Review Board Committee of King Fahad Medical City (IRB number 15-175).However,due to its archival nature or using leftover samples,permission for patient consent was waived.
2.2.DNA extraction from whole blood and paraffin-embedded tissue
DNA extraction from whole blood was performed using MagNa Pure Compact instrument(Roche Diagnostics GmbH,Roche Applied Science,Mannheim,Germany).The ReliaPrep™formalin-fixed,paraffin-embedded gDNA Miniprep System kit (https://worldwide.promega.com) was utilized for DNA extraction from paraffin-embedded tissue block [16,17].Both methods were performed according to the manufacturer’s instructions.Nanodrop® 2000c spectrophotometer (ThermoFisher Scientific,Wilmington,DE,USA)was used to determine DNA quality and quantity,and a DNA concentration of 20 ng/μL with an absorbance ratio of 260 nm/280 nm between 1.6 and 2.0 was accepted for the assays genotyping.
2.3.SNP genotyping
Thirteen SNPs including rs4430796,rs1859962,rs16901979,rs6983267,rs1447295,rs1571801,rs4054823,rs1545985,rs7652331,rs629242,rs13149290,rs251177,and rs10492519 were genotyped.Using kits from Applied Biosystems(Foster City,CA,USA),the SNP assay contains sequence-specific forward and reverse primers.Two TaqMan®minor groove binder polymerase chain reaction (PCR) probes and dyes(VIC™and FAM™,ThermoFisher Scientific,Wilmington,DE,USA) were used to detect alleles 1 and 2 for each SNP,according to manufacturer instructions.The LightCycler®480 instrument (Roche Diagnostics Ltd,Ferrenstrasse,Rotkreus,Switzerland) was used for data collection.
Briefly,in each well of a LightCycler®480 multi-well plate 96,we added 1.25 μL of 1× working SNP genotyping assay,12.5 μL of TaqMan®universal master mix(ThermoFisher Scientific,Wilmington,DE,USA),7.25 μL of DNase-free water,and 4 μL of the 5 ng/μL DNA sample.Then,we sealed the plate,mixed it gently,and centrifuged it briefly.We used LightCycler®480 instrument with 95°C for 10 min,92°C for 15 s,and 60°C for 1 min for each cycle(40 cycles) program.The LightCycler®480 SW 1.5.1 software(https://lightcycler480-software.software.informer.com/1.5)was used for the data collection and analysis for SNP alleles discrimination.Multiple DNA samples were replicated to ensure that a single observation was not spurious.Furthermore,blind testing of coded samples that contained DNA template as positive controls and four wells of no DNA template as negative controls were included in each PCR setup run to ensure the high quality of our laboratory work[18].
2.4.Statistical analysis
Conformation for Hardy-Weinberg equilibrium was analyzed in control data implementing the Chi-squared test distribution (degree of freedom=1) to detect the differences between the expected and observed values in the tested SNPs as described by Rodriguez et al.[19].The frequencies of alleles,genotypes,and the genotype models for each SNP (dominant and recessive traits) were derived by an algorithm based on the direct counting method using the SNP stats software (https://www.snpstats.net/start.htm)and the results were expressed as percentage at 95% confidence interval.Fisher’s exact test was used to compare the difference between patient and control groups.The log (p-value) was used to determine each SNP allele at 5%.The Bonferroni correction test was used whenever possible to adjust multiple comparisons,and the final result was presented as a correctedp-value.
3.Results
3.1.Study population characteristics
The study included 85 patients with PCa (age: range 60.0-70.0 years,mean 69.5 years,median 71.0 years),120 patients with BH (age: range 62.0-76.0 years,mean 67.5 years,median 70.0 years),and 115 healthy normal control(age:range 45.0-67.0 years,mean 52.6 years,median 60.0 years).Clinical information including age demographics was obtained.
3.2.SNPs
We calculated Hardy-Weinberg equilibrium for allele frequencies distribution in healthy controls only to avoid distortion in the case of tested markers being associated with the disease [20].The obtained data (Table 2) have shown a conformation to expected distribution (p>0.01) in 11 SNPs that include rs4430796,rs1859962,rs16901979,rs6983267,rs1571801,rs4054823,rs7652331,rs629242,rs13149290,rs251177,and rs10492519.The rs1447295 and rs1545985 SNPs did not show conformation to expected distribution for Hardy-Weinberg equilibrium (p<0.01).

Table 2 The p-values for Hardy-Weinberg equilibrium for the studied SNPs in healthy control samples.
As shown in Table 3,rs16901979A allele revealed higher significant frequency in PCa patients when compared to healthy controls after applying the Bonferroni correction test(24%vs.8%,odds ratio [OR]=3.6,p=0.0001,correctedp=0.0013).However,rs16901979A allele did not satisfy Bonferroni correction test in the comparison between PCa group against BH group (24%vs.13%,OR=2.0,p=0.008, correctedp=0.104).No other alleles have shown significant differences in the studied groups.
SNP genotypes were analyzed and presented in Table 4.The rs16901979 CA genotype was found at higher significant frequency in PCa patients compared to normal healthy controls after Bonferroni correction test (43%vs.14%,OR=4.6,p=0.0001,correctedp=0.0012).Significant differences were also found in rs16901979 CA genotype but did not satisfy Bonferroni correction test when comparisons were made between BH group and healthy control group(25%vs.14%,OR=2.1,p=0.03,correctedp=0.36),and PCa and BH patients (43%vs.25%,OR=2.2,p=0.009,correctedp=0.117).No other significant associations were found in term of genotype models.
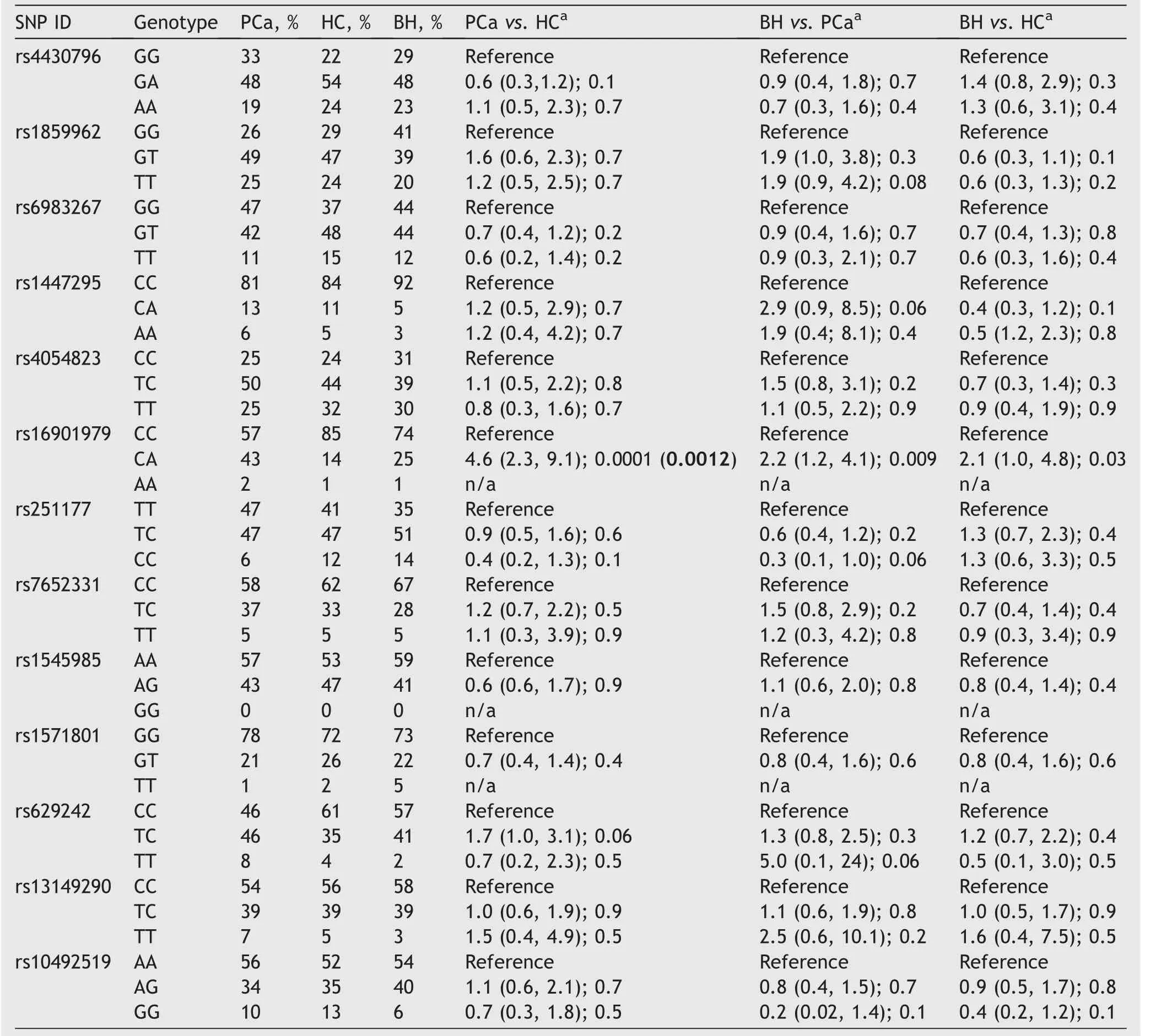
Table 4 Genotype frequencies of studied SNPs in HCs (n=115) and PCa patients (n=85) and BH patients (n=120).
4.Discussion
Over the past decade,genome-wide association studies have identified thousands of SNPs that are associated with disease risk,particularly cancer.The majority of SNPs are known to localize to non-coding regions of the genome,where they are generally thought to map to distal transcriptional enhancers[21].Generally,SNPs may have a functional role in causing amino acid changes,mRNA transcript instability,and transcription factor binding affinity variations [22,23],and therefore,can influence the development of cancer.In this study,we have investigated 13 different SNPs including rs4430796 (TCF2),rs1859962 (CASC17),rs6983267 (CASC8,CCAT2),rs1447295 (CASC8),rs1571801 (DAB2IP),rs1545985(FYCO1),rs7652331(FYCO1),rs29242(KIAA1211),rs13149290(ZNF827),rs251177 (FCHSD1),rs10492519 (FAM124A),rs16901979 (intergenic),and rs4054823(intragenic) in Saudi patients and healthy controls utilizing TaqMan real-time PCR assays.These SNPs have been previously shown to have an association with PCa in some populations.The field of prostate carcinogenesis in affected patients is so difficult in terms of diagnosis and therapeutics;the main reason is that there are no specific biomarkers to differentiate between benign and cancerous conditions[24].Both PCa and BH are identified by enlargement of the prostate gland,but the latter is neither cancerous nor fatal.Some studies suggested that BH may play as a risk factor for PCa with inconsistent evidence[25].
Our results have shown that rs16901979 CA genotype has a significantly higher frequency in PCa patients compared to normal healthy controls after applying the Bonferroni correction test(43%vs.14%,OR=4.6,p=0.0001,correctedp=0.0012),suggesting this genotype has a strong association with PCa development in Saudi males and could be considered a risk factor for this disease.The rs16901979 SNP was found in association with PCa in a case-control study by Robbins et al.[26];the study included 490 PCa patients and 567 healthy controls from African American population.A similar association was found in another study by Chen et al.[27]and in another multi-center study of the Swedish population that included 2893 PCa patients and 1781 control subjects[13].The rs16901979 is considered an intergenic SNP residing within the non-protein-coding region of theCASC8gene located at chromosome 8q24.Genome-wide association studies and several case-control studies have demonstrated significant associations between specific variants in this gene (CASC8) and PCa,suggesting that chromosome 8q24 regions can independently influence the risk for PCa development and advanced disease status [28,29].
Our results also revealed that the rs629242T allele was found at a significantly higher frequency among PCa patients (Table 3).A study investigating the role of rs629242 in PCa has found that rs629242T allele conferred a 29% increased risk for the disease progression in African American population [30].The rs629242 SNP is present within theKIAA1211gene located at the 4q12 region,which encodes an actin cytoskeletal protein regulator to maintain the epithelial cells suppressing tumorigenesis integrity[31].
On the other hand,the rs1447295 SNP did not exhibit significant association with PCa,in concordance with similar findings by Robbins et al.[26]in the African American population study[26].However,the rs1447295 SNP was demonstrated in association with PCa in a meta-analysis study carried out by Cheng et al.[29].It was also identified in association with PCa in Northern Chinese men[32].These contradictions may be attributed to ethnic and geographic differences and may reflect the concept of interaction between genetic susceptibility and surrounding environmental factors that may play an important role in disease development [26].However,based on our findings,this allele and others were found in higher frequency in nodular hyperplasia patients,which may be used as markers in differentiating between benign and malignant prostate tumors in Saudi males.The small sample size is considered a limitation for this study and a replicated multicenter study with larger population is recommended.
5.Conclusion
Our study has highlighted that rs16901979 SNP is associated with PCa development in Saudi males.This finding will benefit researchers in PCa genetic studies and will encourage other studies for more understanding and clarification.
Author contributions
Study concept and design: Awad Elsid Osman,Asim Ali Elbagir.
Data acquisition and analysis: Awad Elsid Osman,Sahar Alharbi.
Drafting of the manuscript:Awad Elsid Osman,Asim Ali Elbagir.
Critical revision of the manuscript: Atif Ali Ahmed.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
This study is fully supported by King Fahad Medical City,Riyadh,Saudi Arabia (IRF No: 017-059).The authors would like to thank the patients and volunteers who participated in this study.
杂志排行
Asian Journal of Urology的其它文章
- Transurethral resection of bladder tumor:A systematic review of simulator-based training courses and curricula
- Etiology and management of urethral calculi:A systematic review of contemporary series
- Oncologic outcomes with and without amniotic membranes in robotic-assisted radical prostatectomy: A propensity score matched analysis
- The risk of prostate cancer on incidental finding of an avid prostate uptake on 2-deoxy-2-[ 18F]fluoro-D-glucose positron emission tomography/computed tomography for non-prostate cancer-related pathology:A single centre retrospective study
- Prevention of thromboembolic events after radical prostatectomy in patients with hereditary thrombophilia due to a factor V Leiden mutation by multidisciplinary coagulation management
- Transurethral prostate surgery in prostate cancer patients: A population-based comparative analysis of complication and mortality rates
