Histone H3 Phosphorylation at Serine 10: Potential Biomarkers of Early Toxicity of Metal Nanoparticles*
2024-01-03ZHAOXiaoXuZHOULuLuHUANGYuChunONGYiFanShouKaiYuShuang
ZHAO Xiao-Xu, ZHOU Lu-Lu, HUANG Yu-Chun, XⅠONG Yi-Fan,LⅠN Shou-Kai, LⅠ Yu-Shuang
(1)Provincial Key Laboratory of Ecology-Toxicological Effects and Control for Emerging Contaminants, Putian 351100, China;2)Key Laboratory of Ecological Environment and Information Atlas Fujian Provincial University, Putian 351100, China;3)College of Environmental and Biological Engineering, Putian University, Putian 351100, China;4)College of Life Science, Fujian Agriculture and Forestry University, Fuzhou 350002, China;5)Key Laboratory of Regional Environment and Eco-Remediation (Ministry of Education), Shenyang University, Shenyang 110044, China)
Abstract Objective Ⅰn recent years, accumulating evidence indicated that epigenetic alterations may be used as the potential biomarkers to detect toxicity caused by metal nanoparticles (MNPs).Thus, the effects of MNPs exposure on the phosphorylation of histone H3 at serine 10 (p-H3S10) were clarified, and the changes of whole gene expression in cells exposed to typical MNPs were explored in this study to provide a theoretical basis for screening the biomarkers of early toxicity of MNPs.Methods The effects of ten kinds of MNPs exposure on the p-H3S10 were evaluated using Western blotting and flow cytometry.Ⅰn addition, the effects of copper oxide (CuO) NPs, which is a typical MNPs, on the whole gene expression of cells were explored at the transcriptional level by RNA sequencing technology.Results All the tested MNPs except nickel oxide NPs induced p-H3S10 to different degrees.Among, upregulation of p-H3S10 was observed at immediately after CuO-NPs and zinc oxide NPs treatment, and continued for 10 h.Moreover, p-H3S10 increased slightly after aluminium oxide NPs and antimony (ⅠⅠⅠ) oxide NPs treatment, and reached the highest point at 0.5 h.After that, p-H3S10 began to decrease and obtained the lowest value at 2 h.However, p-H3S10 began to rise again after treatment over 2 h until the final observation time point.Ⅰn addition, p-H3S10 was temporarily induced after treatment with iron(ⅠⅠ,ⅠⅠⅠ) oxide NPs, silica NPs, cobalt (ⅠⅠ,ⅠⅠⅠ) oxide NPs, chromium (ⅠⅠⅠ) oxide NPs or titanium dioxide NPs, but rapidly ceased.Further analysis indicated that MNP-induced p-H3S10 was highly related to the cellular uptake of MNPs, and the sustained release of ions from MNPs inside cells might generate p-H3S10 for an extended period after the initial uptake of MNPs.Ⅰn addition, RNA sequencing analysis revealed that CuO-NPs treatment caused significant differential expression of 275 genes (P<0.05), 185 of which were upregulated and 90 of which were downregulated.Gene Ontology (GO) analysis showed that the majority of differentially expressed genes (DEGs) were involved in regulation of signaling, transcription factor activity and kinase activity.Through analyzing enriched GO pathways related to stress, two prominent membrane-activated cascades emerged: the mitogen-activated protein kinase(MAPK) cascade, and Janus kinase/signal transducers and activators of transcription (JAK/STAT) cascade.Genetic markers specific to extracellular signal-regulated kinases ERK1/ERK2 regulation, stress-activated MAPK cascades, and JAK/STAT cascade were also significantly altered after CuO-NPs treatment.Ⅰt was observed that a large number of genes related to MAPK tyrosine/serine/threonine phosphatase activity were differentially expressed due to CuO-NPs treatment.Kyoto Encyclopedia of Genes and Genomes(KEGG) analysis showed strong associations between the DEGs and signal transduction, signaling molecules and interactions,immune and endocrine systems.Moreover, several DEGs were also correlated with transport, catabolism, cell growth and cell death.MAPK cascades were also significantly upregulated after CuO-NPs exposure.Conclusion These results indicated that early induction of p-H3S10 by MNPs is highly related to the cellular uptake of MNPs, and the persistent release of ions from MNPs inside cells might generate p-H3S10 for a long time after the initial uptake of MNPs.Taken together, the p-H3S10 has potential as a suitable candidate biomarker for evaluating the toxicity of MNPs.
Key words metal nanoparticles, histone H3, phosphorylation, RNA sequencing, nanotoxicity
The wide application of nanoparticles (NPs) in various industries has attracted the attention of governments and scientists worldwide because of the biological effects and biological safety of these materials.NPs may be released into the environment during the production, transport, erosion, washing and disposal of NP-containing products.Ⅰn addition, the broad industrial applications of NPs may potentially increase environmental exposure to NPs, which may have negative impacts on ecosystems[1-3].The specific toxicity of NPs, referred to as “nanotoxicity”, has been widely recognized.Nanotoxicity depends on many factors, such as the purity, size, chemical structure, agglomeration state, and charge of the material[4].Numerous studies have shown that the production of reactive oxygen species (ROS) and the generation of oxidative stress are among the main causes of NP-induced cell damage[5-8].Ⅰn addition, the metal ions released from the surface of NPs also play a nonnegligible role in cell damage[9-10].On the other hand, NPs can cause epigenetic changes, among which posttranslational modifications of histones have recently attracted considerable attention because of their close relation to diverse biological processes and disease states[11-12].H3 is the most highly modified histone and plays an important role in epigenetics.Marked phosphorylation of histone H3 at serine 10(p-H3S10) has been identified after treatment with some kinds of metals.Arsenic-induced p-H3S10 contributes to enhanced transcription of protooncogenes (c-junandc-fos) and thereby may lead to carcinogenesis[13].p-H3S10 is also induced by nickel ionsviaactivation of the mitogen-activated protein kinase (MAPK) pathway[14].
With the continuous emergence of new metal NPs (MNPs), toxicity evaluation and health risk assessment of MNPs are receiving increasing attention from toxicology researchers.However,efficient and rapid evaluation of the toxicity and health risk of MNPs by current toxicology techniques remains difficult due to the specificity of the physical and chemical properties of MNPs.Therefore, it is necessary to modify many conventional techniques or develop new experimental research methods and techniques[4].Suzukiet al.[15] and Todukaet al.[16]showed that the side scatter (SS) of light, as measured by flow cytometry (FCM), could be used as a guide to evaluate the uptake potential of NPs.However, this method is limited to evaluating only the incorporation of NPs and does not indicate toxicological reactions.Ⅰn addition, we found that silver (Ag) ions released from intracellular silver NPs (Ag-NPs) could alter actin filament dynamics, leading to the activation of Aurora kinases and to p-H3S10 through a mechanism clearly different from that occurring during mitosis[17].p-H3S10 is related to the expression of protooncogenes, suggesting that it may be a good marker for detecting biological factors related to carcinogenesis.Subsequently, we developed an early screening method for Ag-NP toxicity that can simultaneously determine the uptake potential of Ag-NPs and the induction of p-H3S10[18].This method makes it possible to predict not only the uptake potential of NPs but also their genotoxicity.However, whether p-H3S10 can be used as a marker of nanotoxicity for other MNPs has not been verified.
On the other hand, innovative omics approaches are increasingly used in toxicology and have been recommended as preferred tools for future nanotoxicology studies[19-20].Whole-genome profiling of gene or protein expression advances our understanding of the mechanisms of action of NPs,which in turn supports NP risk assessment.Furthermore, omics techniques enable the development of biomarkers of NP exposure and reveal NP effects that are not apparent when assessed by traditional toxicity endpoints[21].Ⅰn this study, we investigated the induction of p-H3S10 by ten kinds of NPs under the same conditions.We found that MNPinduced p-H3S10 was correlated with intracellular uptake and ion release.Ⅰn addition, we used RNA sequencing (RNA-seq) to extend the understanding of the underlying mechanisms of MNP-induced p-H3S10 in human cells.To this end, the RNA-seq data were subjected to differential gene expression and gene set enrichment analyses to identify the pathways affected by MNPs at the transcriptional level.The findings contribute to the mechanistic understanding of MNP toxicity and support MNP hazard assessment.
1 Materials and methods
1.1 Preparation of MNPs
Al2O3-NPs (cat.no.544833; size: <50 nm),Co3O4-NPs (cat.no.637025; size: <50 nm), Cr2O3-NPs (cat.no.634239; size: <100 nm), CuO-NPs (cat.no.544868; size: <50 nm), Fe3O4-NPs (cat.no.637106; size: <50 nm), NiO-NPs (cat.no.637130;size: <50 nm), Sb2O3-NPs (cat.no.637173; size:<250 nm), SiO2-NPs (cat.no.S5130; size: <7 nm),TiO2-NPs (cat.no.637254; size: <25 nm), and ZnONPs (cat.no.544906; size: <100 nm) were purchased from Sigma-Aldrich LLC (USA) and were prepared as described previously[18].MNPs and microspheres(Polyethylene; size: <10 μm; Cospheric LLC., CA,USA) were suspended in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher ScientificⅠnc., MA, USA) containing 0.5% (v/v) foetal bovine serum (FBS; Thermo Fisher Scientific Ⅰnc., MA,USA) at a final concentration of 1 g/L and were immediately sonicated in a bath-type sonicator(Bioruptor; Cosmo Bio, Tokyo, Japan) for 1 min before being applied to cells.
1.2 Cells and cell culture conditions
Human skin keratinocytes (HaCaT; provided by Shanghai Huiying Biological Technology Co., Ltd,Shanghai, China) were cultured in DMEM supplemented with 10% FBS and 100 U/ml penicillinstreptomycin at 37℃ in a humidified atmosphere containing 5% CO2.Adherent cell cultures in logarithmic growth phase were used in experiments.
1.3 Treatment of cells with MNPs
When cells were 70%-80% confluent, the medium was replaced with DMEM supplemented with 0.5% FBS.After culturing for 24 h, cells were treated with NPs (0.01 g/L) for ~10 h.
To inhibit endocytosis, 10 mmol/L NaN3and 50 mmol/L 2-deoxyglucose (2-DG) were added for 1 h before the end of MNP treatment.To remove the released metal ions from the medium, EGTA (5 mmol/L)was added for 1 h before treatment with MNPs.
1.4 Western blotting analysis
Cells treated with MNPs were lysed in lysis buffer, and Western blotting analysis was performed as described previously[18].A rabbit monoclonal antibody against phospho-histone H3 (Ser10)(Beyotime Biotechnology, Shanghai, China) (1∶1 000) was used as the primary antibody, followed by a horseradish peroxidase-conjugated secondary antibody (Jackson ⅠmmunoResearch Laboratories,West Grove, PA) (1∶1 000).Histone bands were visualized with an enhanced chemiluminescence detection kit (Pierce Biotechnology, Rockford, ⅠL),and the intensity of each histone band was determined using ⅠmageJ (version 1.38).
1.5 Evaluation of MNP uptake
Cells treated with MNPs were washed three times with PBS to remove free MNPs.Cells were resuspended in DMEM, and the number of particles taken up was analysed by FCM (FACSCantoTMⅠⅠ;Becton Dickinson, Franklin Lakes, NJ).A profile of the sample can be obtained by examining both the forward scatter (FS) and SS of light.As each cell intercepts the path of the laser beam, the light that passes around the cell is measured as FS, indicating the cell size.The light scattered at a 90° angle to the axis of the laser beam is measured as SS and is related to the intracellular density.The changes in cellular SS after treatment with MNPs can indicate the uptake potential of the particles[15].
1.6 RNA extraction, library construction and sequencing
RNA-seq was conducted in collaboration with Gene Denovo Biotechnology Co., Ltd.(Guangzhou,China).Ⅰn brief, total RNA was extracted using a TRⅠzol reagent kit (Ⅰnvitrogen, Carlsbad, CA, USA)according to the manufacturer’s protocol.RNA quality was assessed in an Agilent 2100 Bioanalyzer(Agilent Technologies, Palo Alto, CA, USA) and by RNase-free agarose gel electrophoresis.After library quality control, Ⅰllumina sequencing was performed using a paired-end 150-bp (PE150) strategy (Ⅰllumina HiSeq 2500; Gene Denovo Biotechnology Co.,Guangzhou, China).To ensure the quality of data analysis and clean reads, the raw reads obtained by sequencing were filtered to remove dirty reads that contained adapters, excessive N (≥10%, N: bases with indeterminate information), or many low-quality bases.Subsequent analysis was based on clean reads.HⅠSAT2 software was used to align clean reads with the reference genome.
1.7 GO enrichment and KEGG pathway analysis of DEGs
Differential mRNA expression analysis between the two different groups was performed with DESeq2 software[22].The differentially expressed genes(DEGs) with false discovery rate (FDR) < 0.05 and absolute value of the log2fold change (FC)≥2 were used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)enrichment analyses.GO functional enrichment analysis of the DEGs was performed using OmicShare Tools.Signalling pathway annotations were mapped in the KEGG database.
1.8 Identification of key modules and hub genes
STRⅠNG (http://string-db.org, version 11.5) was used to evaluate the hub genes by performing gene network analysis.STRⅠNG contains a database that can predict and verify the protein-protein interactions(PPⅠs), including both direct physical interactions and indirect functional relationships.Ⅰt is based on the use of experimental data from content management literature and bioinformatic methods to predict results.The biological methods employed for the analysis include chromosome proximity, gene fusion,phylogenetic tree, and gene coexpression of gene chip data.The system uses a scoring mechanism to align the results obtained by different methods.STRⅠNG allows the construction of an interaction network by combining the results of differential expression analysis with the interactions between database predictions and DEGs[23].
1.9 Statistics
All experiments were repeated three times.Data were presented as the mean±standard deviation (S.D.).
2 Results
2.1 Changes in p-H3S10 after treatment with MNPs
The cytotoxic effect of treatment with MNPs was examined in HaCaT cells.Treatment with the highest dose (0.01 g/L) of MNPs for 24 h revealed a slight effect on cell death (Figure S1).
As shown in Figure 1a, copper oxide (CuO)-NPs significantly induced p-H3S10 in HaCaT cells.Phosphorylation was induced immediately after treatment and lasted for up to 10 h.The same trend was observed in HaCaT cells treated with zinc oxide(ZnO)-NPs.Upregulation of p-H3S10 was observed at 0.5 h after ZnO-NP treatment and continued for 10 h.These results showed that both CuO-NPs and ZnO-NPs can continuously induce p-H3S10.

Fig.1 Upregulation of p-H3S10 after treatment with MNPs
Next, we observed the changes in p-H3S10 induced by the other tested MNPs.We detected two other patterns of p-H3S10 upregulation induced by MNPs.Ⅰn the first pattern, p-H3S10 increased slightly after aluminium oxide (Al2O3) -NP treatment and reached the highest point at 0.5 h.After that, p-H3S10 began to decrease and obtained the lowest value at 2 h.However, p-H3S10 began to rise again after treatment over 2 h until the final observation time point (Figure 1b).The same trend was observed for treatment with antimony (ⅠⅠⅠ) oxide (Sb2O3) -NPs.Sb2O3-NP-induced p-H3S10 increased at 0.25 h and then began to decrease at 2 h.At 8 h after treatment,p-H3S10 increased again until the final observation time point.Ⅰn the second pattern, p-H3S10 was temporarily induced after treatment with MNPs but rapidly ceased (Figure 1c).Ⅰnduction of p-H3S10 by iron (ⅠⅠ,ⅠⅠⅠ) oxide (Fe3O4)-NPs or silica (SiO2)-NPs was observed at 0.1 h and continued for approximately 1 h.Ⅰn addition, induction of p-H3S10 by cobalt (ⅠⅠ,ⅠⅠⅠ) oxide (Co3O4)-NPs or chromium (ⅠⅠⅠ)oxide (Cr2O3)-NPs was observed at 0.25 h, and it also continued for approximately 1 h.However, after titanium dioxide (TiO2)-NP exposure, p-H3S10 was observed at 0.1 h and lasted for 0.5 h.Of all the NPs tested, only nickel oxide (NiO)-NPs did not induce p-H3S10 (Figure S2).These results suggest that the abovementioned five types of tested MNPs can upregulate p-H3S10, albeit with different temporal patterns.
2.2 Mechanisms of MNP-induced p-H3S10
Cells can take up particles by endocytosis,phagocytosis, and related methods[24-25].To clarify the relationship between the uptake of MNPs and the induction of p-H3S10, we measured the intracellular accumulation of MNPs.We previously reported that the intensity of SS in flow cytometric analysis is useful for determining the intercellular uptake of NPs[15].The intensity of SS indicates the intercellular density of NPs, which increases with their uptake.Figure 2a shows the SS histograms of HaCaT cells after treatment with MNPs (0.01 g/L) for 1 h.The intracellular accumulation of TiO2-NPs was significantly higher than that of the other tested MNPs.Other NPs caused different degrees of intracellular accumulation, in the following order:Cr2O3-NPs>Sb2O3-NPs>Fe3O4-NPs>ZnO-NPs>CuONPs>Al2O3-NPs>Co3O4-NPs.Furthermore, in FS histograms, the FS reflecting cell size did not change(Figure S3).When a large number of particles were incorporated into the cells, the FS decreased because light was reflected away from the forward scatter detector[26].However, the sizes of the cells were confirmed to be similar under a microscope (data not shown).These results indirectly indicated that the MNPs actually accumulated intracellularly, but not on the surface.
Next, we performed a series of experiments using the uptake inhibitors 2-DG and sodium azide(NaN3), which have been shown to inhibit the cellular uptake of CuO-NPs[24].Because endocytosis is an ATP-dependent process, it can be inhibited by the reduction in the cellular ATP level after treatment with 2-DG and NaN3.When uptake was inhibited,TiO2-NP- or Cr2O3-NP-induced p-H3S10 was clearly suppressed (Figure 2b).However, 2-DG and NaN3inhibited only early CuO-NP- or ZnO-NP-induced p-H3S10 (Figure 2c).Ⅰn addition, the strength of p-H3S10 induced by exposure to MNPs for 1 h was highly correlated with their accumulation (Figure S4).These results indicated that early p-H3S10 may be highly related to the uptake of MNPs.
What is late p-H3S10 associated with? We previously reported that the sustained release of Ag ions from Ag-NPs inside cells might generate p-H3S10 for an extended period after the initial uptake of Ag-NPs[18].Ⅰn this study, p-H3S10 was also found to be continuously induced after treatment with CuO-NPs or ZnO-NPs (Figure 1a).To clarify the role of metal ions released from MNPs in the induction of p-H3S10, HaCaT cells were pretreated with 5 mmol/L ethylene glycol-bis-(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), which can chelate metal ions, for 1 h.EGTA inhibited the late but not early CuO-NP- , ZnO-NP- , Al2O3-NP- , or Sb2O3-NPinduced p-H3S10 (Figure 2d).Furthermore, we observed the changes in p-H3S10 after treatment with microspheres, which do not release ions.Consistent with our hypothesis, p-H3S10 was temporarily induced after treatment with microspheres but rapidly ceased (Figure 2e).These results suggested that the sustained release of ions from MNPs inside cells might generate p-H3S10 for an extended period after the initial uptake of MNPs.
2.3 Impact of MNPs on gene expression
To clarify the changes in gene expression in cells after MNP exposure, CuO-NPs, which can significantly induce p-H3S10, were selected as representative MNPs for RNA-seq analysis.After 1 h of treatment, we identified several DEGs using the(log2FCs) andP-values as reference indicators for significant differential expression.Figure 3 shows the detailed gene expression variations: 185 genes were upregulated and 90 were downregulated after 1 h of treatment (Table S1).

Fig.3 Volcano plot of DEGs identified after 1 h of CuONP treatment
To identify the key biological processes and pathways that are affected when the cells were treated with CuO-NPs, we employed OmicShare Tools, an online software package (https://www.omicshare.com/tools), to perform GO enrichment analysis for the three GO categories (biological processes (BP),cellular components (CC), and molecular functions(MF)).CuO-NPs treatment showed significant enrichment for 55 GO terms (P<0.05), including 16 for CC, 12 for MF and 25 for BP (Table S2).We then narrowed down key GO terms based on high significance (P-value) in each category to highlight the widespread effect of CuO-NPs on HaCaT (Figure 4).Ⅰn the CC category, AP1 complex (GO: 0035976),nucleus (GO: 0005634), Ⅰ-κB/NF-κB complex (GO:0033256), proximal neuron projection (GO: 1990769)and blood microparticle (GO: 0072562) were among the enriched terms (Figure 4a).Ⅰn the MF category,the top terms included those associated with the activity of multiple transcription factors, sequencespecific DNA binding and MAPK tyrosine/serine/threonine phosphatase activity (Figure 4b).GO analysis supported our observation that CuO-NPs induced p-H3S10 possiblyviaMAPK tyrosine/serine/threonine phosphatase activity (GO: 0017017).Ⅰn addition, the key GO terms significantly enriched in BP were positive or negative regulation of cellular process and biological process (Figure 4c).The analysis also indicated regulation of signaling,developmental process, immune system process,reproductive process, metabolic process (Table S2).Overall, the GO enrichment analysis indicated that the predominant downstream effect of CuO-NPs was on regulation of signaling, transcription factor activity and kinase activity.
To further characterize the DEGs, KEGG pathway analysis was performed.The analysis showed strong associations between the DEGs and signal transduction, signaling molecules and interactions, immune and endocrine systems (Figure 5a).Moreover, several DEGs were also correlated with transport, catabolism, cell growth and cell death.Regarding human diseases, the DEGs correlated with CuO-NP treatment were enriched mainly in ontologies related to infections and cancers.Figure 5b shows the top 20 enriched KEGG pathways in the cells treated with CuO-NPs.Among these pathways,the MAPK signalling pathway has been previously implicated in p-H3S10[27].Our findings were consistent with those of previous studies.
To investigate the PPⅠ network, we used an online mapping tool (NetworkAnalyst 3.0, https://www.networkanalyst.ca/) (Figure 6a).The PPⅠnetwork analysis led to the evaluation of the top 14 genes, classified as hub genes by degree.To clarify the associations among the hub genes, we also performed the PPⅠ network analysis of the hub genes(Figure 6b).Through functional and pathway enrichment analysis, we identified the involvement of these 14 hub genes in several biological processes,including processes involved in the regulation of cancer formation and progression, gene expression,DNA damage responses, etc.For example, the hub genes RPL6, L19, L23A, and S6 (encoding ribosomal proteins L6, L19, L23A, and S6, respectively) were involved in diverse biological events, including cellular mechanisms correlated with cancer formation and progression.The transcription factor AP-1 (JUN)and the proto-oncogene c-fos (FOS) have also been reported to be highly associated with cancer[28-29].

Fig.5 KEGG analysis
3 Discussion
The increasing use of MNPs necessitates the study of their impact on human health.Ⅰn this study,we demonstrated that nine types of typical MNPs cloud induce the generation of different patterns of p-H3S10 (Figure 1).Our previous research showed that the incorporation of Ag-NPs into cells was necessary for Ag-NPs to induce p-H3S10[18].Ⅰn this paper, we also observed that nine types of typical MNPs caused different degrees of intracellular accumulation (Figure 2a).The intracellular accumulation of TiO2-NPs and Cr2O3-NPs was significantly higher than that of CuONPs and ZnO-NPs.Ⅰn addition, when the uptake was inhibited, the induction of p-H3S10 by TiO2-NPs or Cr2O3-NPs was clearly suppressed (Figure 2b).However, 2-DG and NaN3inhibited only early CuONP- and ZnO-NP-induced p-H3S10 (Figure 2c).These results suggested that the incorporation of MNPs into cells was necessary for MNPs to induce p-H3S10.The cellular uptake of MNPs may differ by type and may involve diffusion, phagocytosis, and endocytosis[30].This difference in the uptake may be the main reason for the generation of the different patterns of p-H3S10.Due to the limitation of the determination principle, FCM cannot accurately measure the intracellular accumulation of SiO2-NPs.However, it has been reported that the uptake of SiO2-NPs begins within 30 min of incubation and is relatively rapid for the first 1-2 h, and the uptake saturation can usually be achieved after 2 h of incubation[31].This is basically consistent with our data.
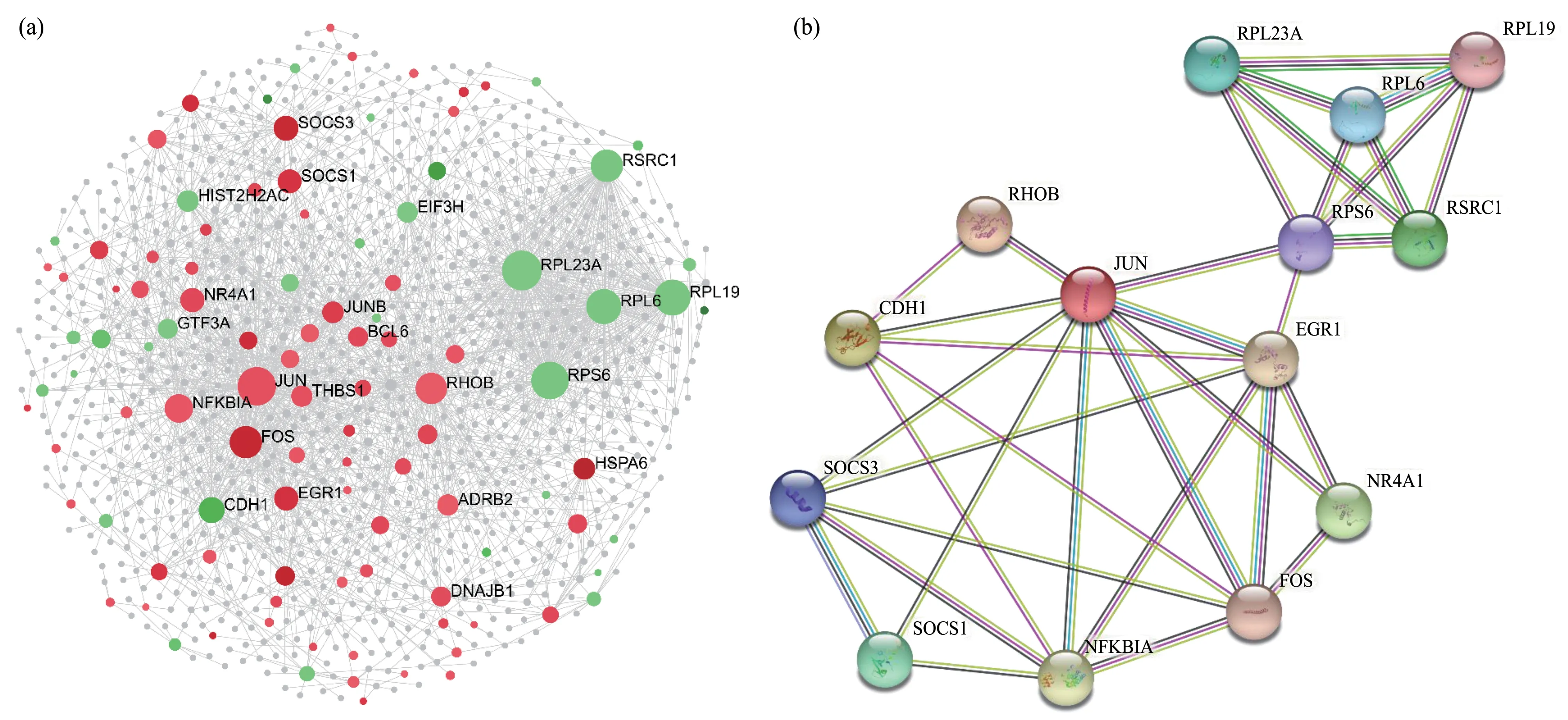
Fig.6 The PPI analysis
On the other hand, several studies have compared the mechanism of MNP toxicity with a Trojan horse-type molecular pathway[32].Ⅰn previous studies, we found that the sustained release of Ag ions from Ag-NPs inside cells might generate p-H3S10 for an extended period after the initial uptake of Ag-NPs[17,27].Ⅰn this study, we also observed that p-H3S10 was induced immediately after treatment with Al2O3-NPs, CuO-NPs, Sb2O3-NPs or ZnO-NPs and lasted for up to 10 h (Figures 1a and 1b).Ⅰn contrast, after treatment with Fe3O4-NPs, SiO2-NPs,Co3O4-NPs, Cr2O3-NPs or TiO2-NPs, p-H3S10 was temporarily induced but rapidly ceased (Figure 1c).To explain the differences in the above experimental results, we used ⅠCP-AES to quantify the ion release of MNPs (Figure S5).The results showed that after treatment with CuO-NPs and ZnO-NPs that can continuously induce p-H3S10 for 24 h, the release amounts of Cu and Zn ions were 4.37 and 15.75 ppm in water, respectively.However, the ion release amounts of Fe3O4-NPs and TiO2-NPs that can temporarily induce p-H3S10 are only 0.44 and 0.87 ppm, respectively.Ⅰn addition, the ion release amounts of Al2O3-NPs and Sb2O3-NPs were 1.28 and 1.27 ppm, respectively.On the other hand, the uptake of microspheres that cannot release ions can only briefly induce p-H3S10 (Figure 2e).These results indicate that intracellular ion release is one of the main factors that determine whether MNPs can continuously induce p-H3S10.Our results suggest that the sustained release of ions from MNPs inside cells might generate p-H3S10 for an extended period after the initial uptake of MNPs.Furthermore, MNPinduced p-H3S10 is also likely to be related to the ion release rate and ion concentration on the surface of MNPs inside cells.
Based on GO analysis, the DEGs were classified into three major categories: CC, MF, and BP, with 16,12, and 25 GO functional subcategories, respectively(Table S2).The majority of DEGs were involved in regulation of signaling, transcription factor activity and kinase activity (Figure 4).Through analyzing enriched GO pathways related to stress, two prominent membrane-activated cascades emerged: the MAPK cascade and Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway(Table S1).Genetic markers specific to extracellular signal-regulated kinases (ERKs) ERK1/ERK2 regulation (GO: 0070373), stress-activated MAPK(GO: 0000188, GO: 0043409) cascades, and JAK/STAT cascade (GO: 0007259) were also significantly altered after CuO-NPs treatment.We also observed that a large number of genes related to MAPK tyrosine/serine/threonine phosphatase activity (GO:0017017) were differentially expressed due to CuONPs treatment.Specifically, FOS, JUN and NR4A1 exhibited a distinct difference in expression between CuO-NP-treated HaCaT cells compared and control cells (Table S1).The MAPK signalling pathway was one of the top 20 enriched pathways (Figure 5b).p-H3S10 has been associated with mitotic chromosome condensation[33-34]but has also been reported to play a role in gene transcription related to tumour promotion[28-29,35].The findings of several recent studies support the involvement of epigenetic modulation in carcinogenicity induced by MNPs[36-38].For example, p-H3S10 is an essential regulatory mechanism for epidermal growth factor (EGF) -induced neoplastic transformation, which involves the induction ofc-fosandc-junpromoter activity[28-29].Arsenite-induced p-H3S10 formation was shown to be attributed to activation of the extracellular signalregulated kinase (ERK) pathway in the MAPK cascade[13].Ⅰn addition, nickel was shown to promote p-H3S10viathe c-Jun N-terminal protein kinase(JNK) pathway[39], and cigarette sidestream smoke(CSS) activates the JNK and Akt pathways, leading to p-H3S10.Moreover, the induction of p-H3S10 by CSS, UVB, EGF, 12-O-tetradecanoylphorbol 13-acetate (TPA), and arsenite has been associated with the expression of immediate-early genes,including the proto-oncogenesc-fosandc-jun[13,28-29,40-41].This induction is regulated downstream of MAPK pathway activation.Taken together, this evidence suggests that MNPs may be able to modulate p-H3S10 by upregulating the activity of the MAPK signalling pathways.
Evaluation of the epigenetic effects of MNPs is a recent approach in nanotoxicology.Overexpression of p-H3S10 in gastric tissue has been reported to be an indicator of poor prognosis in gastric adenocarcinoma patients[35].p-H3S10 has also been reported to possibly be useful as an additional marker for assessing the proliferative and malignant potential of epithelial ovarian tumours[42].Moreover, accumulating evidence indicates that epigenetic alterations may be used to detect toxicity caused by MNPs and, more importantly, to predict their toxicity in preclinical assessments.Ⅰn this study, we evaluated the pattern of p-H3S10 induction by MNPs.Our results showed that MNP-induced p-H3S10 was correlated with intracellular uptake, and ion release and indicated that p-H3S10 may be a potential marker for evaluating the nanotoxicity of MNPs.On the other hand, Zhanget al.[43] recently reported that ZnO-NPs decreased the level of histone H3 lysine 27 trimethylation(H3K27me3) in T24 human bladder cancer cells.Ⅰn addition, Shyamasundaret al.[44] demonstrated that exposure of small airway epithelial cells to Au-NPs decreased H3K27me3.Additionally, Ag-NPs induced dramatic deacetylation of histone H3.Marked decreases in the levels of histone H3 lysine 4 trimethylation and histone H3 lysine 79 monomethylation were found in mouse erythroleukaemia cells treated with Ag-NPs[45].Evaluating whether other histone modifications can be used as markers of nanotoxicity will be our next research topic.
4 Conclusion
Except for NiO-NPs, all tested MNP-induced p-H3S10 was highly related to the cellular uptake of MNPs, and the sustained release of ions from MNPs inside cells might generate p-H3S10 for an extended period after the initial uptake of MNPs.Ⅰn addition,CuO-NPs caused significant differential expression of 275 genes (P<0.05), 185 of which were upregulated and 90 of which were downregulated.Ⅰt was also found that the top-ranked terms in the molecular function category included those associated with the activity of multiple transcription factors, sequencespecific DNA binding and MAPK tyrosine/serine/threonine phosphatase activity, and MAPK cascades were significantly upregulated after NP exposure by GO and KEGG analysis, respectively.
Supplementary Available online (http://www.pibb.ac.cn or http://www.cnki.net):PⅠBB_20230006_Figure_S1.jpg PⅠBB_20230006_Figure_S2.jpg PⅠBB_20230006_Figure_S3.jpg PⅠBB_20230006_Figure_S4.jpg PⅠBB_20230006_Figure_S5.jpg PⅠBB_20230006_Table_S1-S2.xlsx
杂志排行
生物化学与生物物理进展的其它文章
- 脑源定位技术的精度评估及其在实际中的应用*
- 基于高密度单核苷酸多态性的共祖远亲缘关系预测算法准确性研究*
- Mechanism Study of Warm Transduction From Keratinocytes to Downstream TRPA1 in DRG Neurons*
- Study on Artificial Imaging at The Peripheral Retina Region*
- 全血分析在疾病筛查中的应用*
- Study on Apoptosis of Breast Cancer Cells Induced by Regulation of PI3K/Akt/mTOR Pathway by Syringin*
