The antifungal properties of terpenoids from the endophytic fungus Bipolaris eleusines
2023-12-29YinZhongFanChunTianShunYaoTongQingLiuFanXuBaoBaoShiHongLianAiandJiKaiLiu
Yin-Zhong Fan ,Chun Tian ,Shun-Yao Tong ,Qing Liu ,Fan Xu ,Bao-Bao Shi* ,Hong-Lian Ai*and Ji-Kai Liu*
Abstract A series of terpenoids (1-17),comprising six new compounds designated bipolariterpenes A-F (1-6) and eleven recognized compounds (7-17),were isolated from the wheat culture of the potato endophytic fungus Bipolaris eleusines.Their structures and stereochemistry were clarified by HRESIMS,NMR,DP4+probability analyses,and computations for electronic circular dichroism (ECD).All compounds are made up of six meroterpenoids,four sesterterpenes and seven sesquiterpenes.Among them,four sesterterpenes (4,5,10,11) were investigated for their antifungal,antibacterial and cytotoxic properties,and six meroterpenoids (1-3,7-9) were evaluated for their antifungal properties.The compounds 7,9,and 10 had substantial antifungal activity against Epidermophyton floccosum at a concentration of 100 μM.No antibacterial and cytotoxic activities were observed.
Keywords Endophytic fungus,Bipolaris eleusines,Terpenoids,Isolation and structure elucidation,Antifungal activity
1 Introduction
Terpenoids,possessed several hundred initial terpene scaffolds,are very important secondary metabolites [1].Terpenoids have a basic unifying trait that all terpenoids are derived from the five carbon precursor molecules dimethylallyl diphosphate and isopentenyl diphosphate[2].According to their chemical skeletons,terpenoids are further categorized into monoterpenes,sesquiterpenes,diterpenes,sesterterpenes and triterpenes as well as meroterpenoids derived from the hybridization of polyketide and terpenoid [3].Terpenoids offer a wide spectrum of biological functions,such as the cancerfighting monoterpene carvacrol [4],the sesquiterpene atractylenolide V with anti-inflammatory effects [5],and the diterpene 7-deoxynimbidiol with antioxidant activity [6].One of the most important sources of terpenoids is endophytic fungi,which have been widely reported to have specific biosynthetic capabilities [7,8].As a result,the metabolites of endophytic fungi deserve to be further investigated.
Bipolarisis a genus of fungi belonging to the family Pleosporaceae [9,14],and many metabolites have been reported from several species of this genus,such as terpenes,polyketides,quinones,peptides and alkaloids [10,11,14].The majority of them have a wide spectrum of beneficial biological qualities,such as antifungal,antibacterial,and cytotoxic properties [12-14].In our previous study onB.eleusines,a series of meroterpenoids,sesquiterpenes and chromones have been characterized [14-16].Among them,bipolarithizole A,a new phenylthiazole-sativene merosesquiterpenoid having a MIC value of 16 μg/mL,inhibitedRhizoctoniasolani[14].To find more potent biological agents,a chemical study onB.eleusineswith a large-scale fermentation was carried out.As a result,seventeen terpenoids,including six undescribed compounds,were isolated from cultures of the fungusB.eleusines.The details of its isolation,structural elucidation,and antifungal,antibacterial as well as cytotoxic activities are reported in this paper (Fig.1).
2 Results and discussion
Compound1was purified as a colorless solid.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C29H41NO6S(10 indices of hydrogen deficiency).The1H NMR spectrum of1(Table 1) revealed indications for seven methyl groups [δH0.85 (t,J=7.4 Hz),1.20 (s),1.19 (s),1.11 (s),1.36 (s),1.44 (d,J=7.1 Hz) and 0.76 (d,J=6.6 Hz)],two oxymethine protons [δH3.29 (m),and 3.26 (m)],as well as a number of aliphatic methylene multiplets.In connection with the DEPT experiment,the13C NMR spectrum (Table 1) revealed 29 carbon resonances traceable to seven methyls,five methylenes,seven methines and ten quaternary carbons.The one-dimensional (1D)NMR data of1were quite similar to those of known substance 19-dehydroxyl-3-epi-arthripenoid A [17],which is a meroterpenoid with sesquiterpenoid and polyketide components.This was followed by more research into the HMBC and1H -1H COSY spectra,which confirmed the planar structure of1.The sesquiterpenoid unit(C-12-C-26) was calculated from the1H-1H COSY correlations of H2-19/H2-20/H-21,H2-15/H2-16/H-17 and H2-12/H-13 as well as HMBC cross-peaks from H3-23 to C-21,C-22,and C-24,from H3-26 to C-13,C-14,and C-15,and from H2-12 to C-13 and C-14 (Fig.2).The polyketide unit (C-1-C-11,C-27 and C-28) was clarified using the1H-1H COSY correlations of H3-1/H2-2/H-3,as well as HMBC cross-peaks from H3-28 to C-2,C-3,and C-4 and from H-27 to C-4,C-5,and C-6(Fig.2).Following that,the two units were linked using the HMBC cross-peaks from H2-12 to C-8,C-9,C-10,C-13,C-14 and C-18.Furthermore,the ROESY correlations of H-21 (δH3.29)/H-19α(δH1.49)/H-17 (δH3.26)/H-13 (δH2.00) and H-19β(δH2.68)/H3-25(δH1.11)/H3-26 (δH1.36)/H-12 (δH5.49) proposed that H-21,H-17,and H-13 wereα-oriented,whereas H3-26,H3-25 and H-12 wereβ-oriented in the sesquiterpenoid section(Fig.3).Based on prior discoveries from single crystal X-ray diffraction and the stereoselectivity of enzymes in biosynthesis,the relative configuration of C-5 is known to be fixed (5S-configuration) [18,19].The absolute configuration of C-3 can be determined using the diagnostic chemical shift of C-2.The13C chemical shift values at the C-2 location are substantially downshifted (δC26.8 for 3SandδC23.6 for 3R),as reported for the 3S-conformation [18].Therefore,the chemical shifts of C-2 atδC27.3 in1revealed that C-3 isS-configured.The proposed structure of1was further confirmed by the quantum chemical calculations of the NMR data (qccNMR) of two diastereoisomers (3S-1and 3R-1).The two epimers were submitted to a thorough conformational screening approach,and the NMR chemical shifts were estimated using the PCM solvent model in methanol at the mPW1PW91/6-31+G(d,p)//M06-2X/def2-SVP level of theory.As a result,the calculated13C NMR data for 3S-1agreed well with its experimental values and DP4+analysis also identified 3S-1as the most instructive structure of1with 100% DP4+chance for all data (Fig.4 and Additional file 1: Fig.S78).Therefore,compound1was defined as (3S,5S,12S,13S,14R,17R,18R,21R)-1.
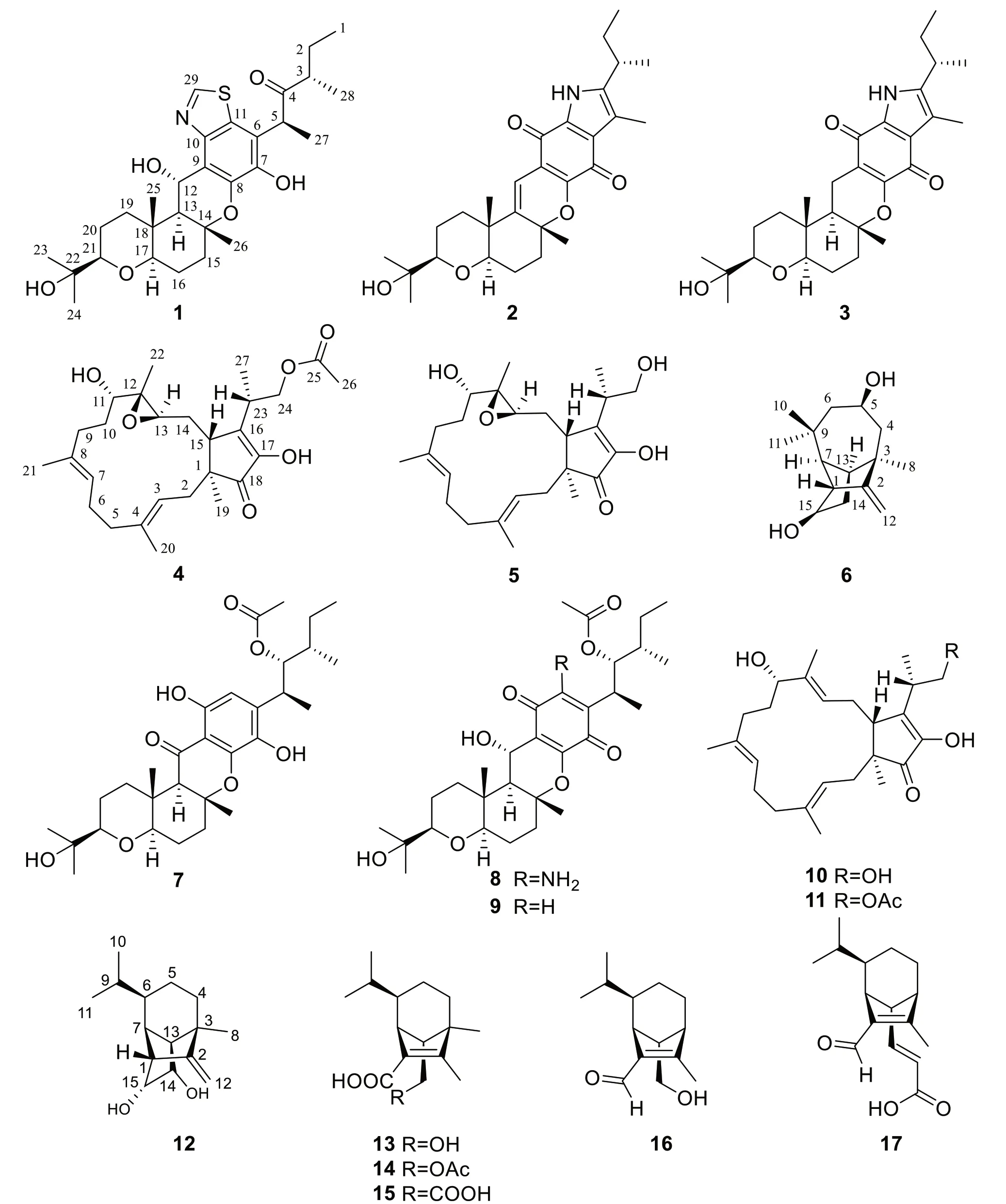
Fig. 1 The chemical structures of compounds 1-17
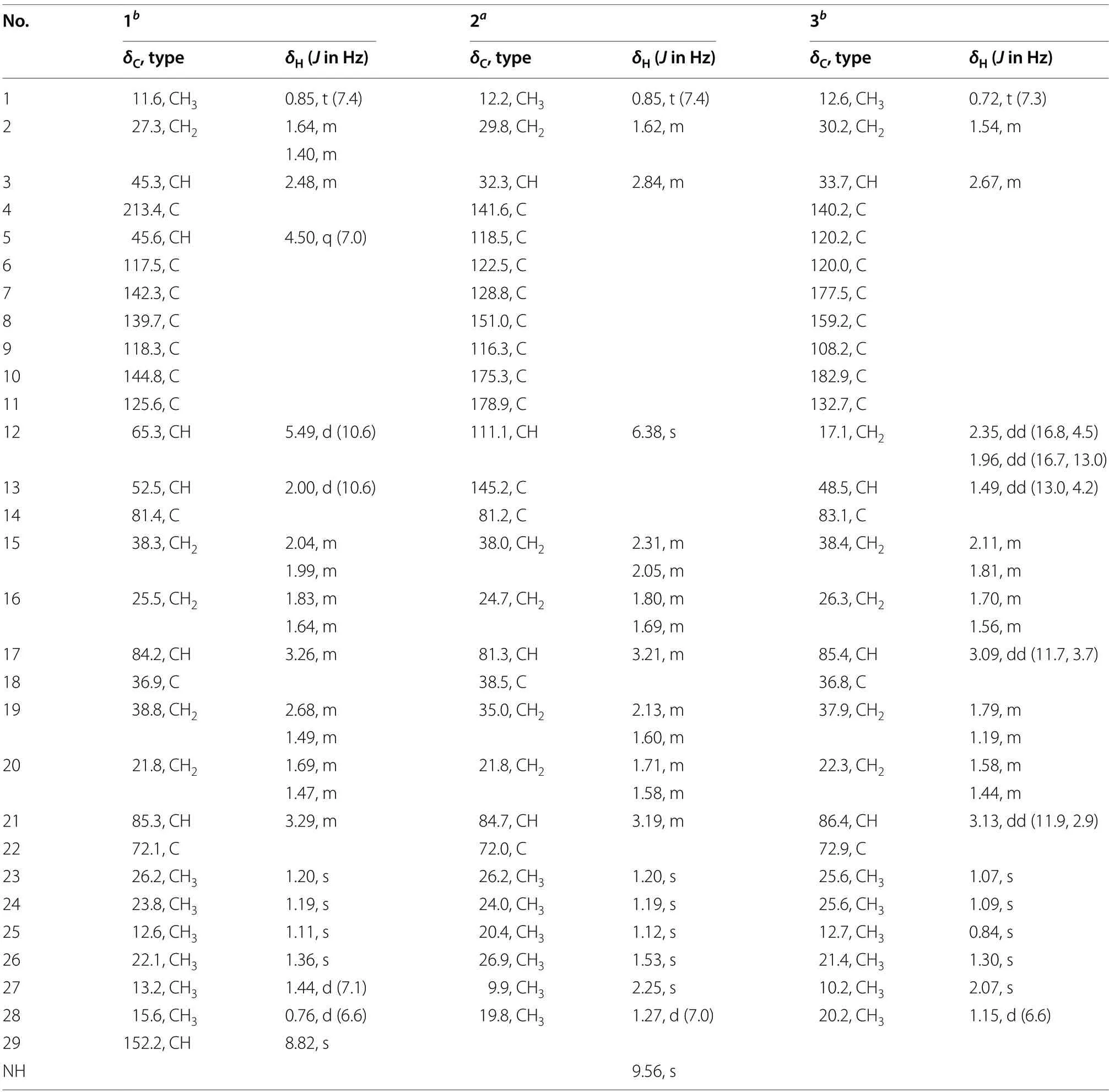
Table 1 1H (600 MHz) and 13C (150 MHz) NMR data of compounds 1-3
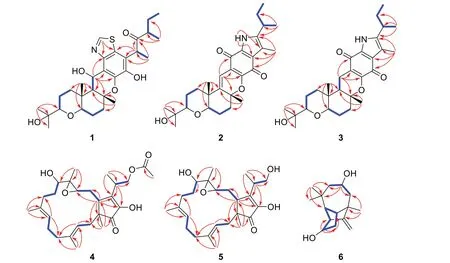
Fig. 2 1H-1H COSY (blue bold lines) and HMBC (red arrows) correlations of compounds 1-6
Compound2was purified as a purple solid.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C28H37NO5(11 indices of hydrogen deficiency).The1H NMR spectrum of2(Table 1) revealed indications for seven methyl groups [δH0.85 (t,J=7.4 Hz),1.20 (s),1.19 (s),1.12 (s),1.53 (s),2.25 (s) and 1.27 (d,J=7.0 Hz)],two oxymethine protons [δH3.21 (m) and 3.19 (m)],as well as a number of aliphatic methylene multiplets.In connection with the DEPT experiment,the13C NMR spectrum (Table 1) revealed 28 carbon resonances traceable to seven methyls,five methylenes,four methines (two oxygenated),and twelve quaternary carbons (three oxygenated carbons and two carbonyls).The1H and13C NMR data of2were structurally similar to those of cochlioquinone G [17] except for the absence of a hydroxyl group at C-12 and the presence of an additional double bond atδC111.1 (C-12) and 145.2(C-13).This hypothesiswas validated by HMBC correlations from H-12 (δH6.38) to C-10 (δC175.3),C-9(δC116.3),C-8 (δC151.0) and C-14 (δC81.2) (Fig.2).The ROESY correlations of H-21/H-17 indicated that H-21 and H-17 were on the same face and were given asα-orientation (Fig.3).The correlations of CH3-25 and CH3-26 established theβ-orientation of H3-25 and H3-26,which was similar to those of cochlioquinone G.Similar to1,the absolute configuration of C-3 was calculated utilizing chemical shifts of C-2 (δC29.8) and the quantum chemical calculations of the NMR data as well as DP4+analysis (Fig.4 and Additional file 1: Fig.S79).Therefore,the absolute configuration of2was defined as (3S,14R,17R,18R,21R)-2.

Fig. 3 ROESY correlations of compounds 1-6
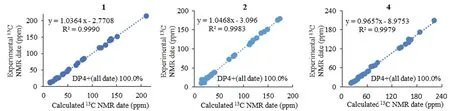
Fig. 4 Correlation plots of experimental and calculated 13C NMR for 1,2,4 and DP4+results of all NMR data
Compound3was purified as a yellow oil.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C28H39NO5(10 indices of hydrogen deficiency).The NMR data (Table 1)showed that compound3was comparable to compound2except for the lack of a double bond group at C-13(δC145.2) and C-12 (δC111.1) and the presence of two alkyl carbons of C-13 (δC48.5) and C-12 (δC17.1).This difference indicated that the double bond of C-13 and C-12 were replaced by a single bond,which was confirmed by HMBC correlations from H-12 (δH2.35) to C-10 (δC182.9),C-8 (δC159.2) and C-14 (δC83.1) and1H-1H COSY correlations of H-13/H-12 (Fig.2).Based on the ROESY profile (Fig.3) and its likely biosynthesis origin from2,the conformation of3was determined as the 3S,13R,14R,17R,18R,21Rconformation,which should be the same as2.
Compound4was purified as a colorless powder.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C27H40O6(8 indices of hydrogen deficiency).Its13C NMR spectra indicated 27 signals generated by six methyl carbons,sevensp3methylene carbons,foursp3tertiary carbons,twosp2methine carbons,twosp3quaternary carbons,foursp2nonprotonated carbons and two carbonyl carbons.The1H and13C NMR data of4(Table 2) weresimilar to those of terpestacin (11) [20].The obvious differences disclosed were that the double bond between C-12 and C-13 was absent and two more oxygenated resonances (δC65.1 and 63.5) occurred,suggesting that the double bond in11is oxidized to form an epoxy ring in compound4,which was confirmed by the1H-1H COSY correlations of H-15/H-14/H-13 and HMBC correlations from H-13 to C-22,C-15 and C-11 (Fig.2).ROESY correlations between H-3/H-5,H-20/H-2,H-7/H-9 and H-21/H-6 show that the two double bonds in4wereE-configured.While the ROESY correlation between H-15/H-23 implied that they were on the identical side and assigned asβ-orientation,the correlation between H-19/H-13,H-19/H-14b,H-14b/H-22,suggested that they were assigned asα-orientation (Fig.3).Furthermore,quantum chemical calculations of the nuclear magnetic resonance data (qccNMR) of two diastereoisomers,(12S,13S)-4and (12R,13R)-4,were undertaken to identify the configuration of the epoxy ring on C-12 and C-13.The two epimers were rigorously conformationally screened,and NMR chemical shifts were estimated in methanol using the mPW1PW91/6-31+G(d,p)//M06-2X/def2-SVP level of theory and the PCM solvent model.With a DP4+probability of 100%,the DP4+analysis also revealed (12R,13R)-4as the most plausible structure for4.(all data) (Fig.4),which was compatible with the ROESY study results presented above.Therefore,the absolute configuration of4was specified as (1S,11S,12R,13R,15R,23S)-4.

Table 2 1H (600 MHz) and 13C (150 MHz) NMR data of compounds 4-6
Compound5was purified as a colorless powder.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C25H38O5(7 indices of hydrogen deficiency).Its13C NMR spectra revealed 25 signals from five methyl carbons,sevensp3methylene carbons,foursp3tertiary carbons,twosp2methine carbons,twosp3quaternary carbons,foursp2nonprotonated carbons and a carbonyl carbon.The NMR spectra of5indicated that it is closely linked to4,except that4lacks an acetyl group and has an additional hydroxyl group at C-24,which is corroborated by the 42 Da drop in molecular weight of5when compared to4.Based on ROESY profile (Fig.3) and its potential biosynthetic origin from4,the configuration of5would be equivalent to4for (1S,11S,12R,13R,15R,23S)-5.
Compound6was purified as a colorless powder.The protonated molecule signal in the HRESIMS was used to calculate its chemical formula,which was C15H24O2(4 indices of hydrogen deficiency).The1H and13C NMR spectra (Table 2) revealed 15 signals from three methyl carbons,threesp3methylene carbons,onesp2methylene carbon,fivesp3tertiary carbon,twosp3quaternary carbons,and onesp2nonprotonated carbon.According to the preceding data,the structural properties of6were comparable to those of longi-β-nozigiku alcohol[21],with the exception of an extra hydroxyl group in6.Chemical shifts at the C-5 (δC67.2) level have increased relative,together with the critical HMBC correlation from H-5 (δH3.87) to C-3 and C-9,suggesting the hydroxyl group was annexed to C-5 (Fig.2).The ROESY correlations of H-15/H-7and H-13/H-7 indicated that H-15,H-13 and H-7 are on the identical side and belong to theα-orientation (Fig.3).The correlations of H-1/H-10 determined theβ-orientation of H-1.Meanwhile,the correlations of H-5/H-11,H-11/H-13 and H-8/H-5 indicated that H-13,H-11,H-8 and H-5 are on the identical side and belong to theα-orientation (Fig.3).Hence,the hydroxyl groups that exist at C-5 isβ-configuration.The absolute configurations of6was determined by comparing the experimental ECD spectra with calculated ECD spectra (Fig.5).Therefore,the absolute configuration of6was defined as (1R,3R,5R,7S,13S,15R)-6.
Eleven known compounds were identified as isocochlioquinone A (7) [11],cochlioquinone N (8) [22],stemphone (9) [23],(-)-terpestacin (10) [24],fusaproliferin(11) [20],cis-sativenediol (12) [25],helminthosporic acid(13),cochliobolin B (14) and cochliobolin A (15) [26],helminthosporol (16) [27] and bipolarisorokin H (17)[28] respectively,based on spectroscopic data comparisons with reported values.
Since terpenoids are known to exhibit a variety of biological activities,the four sesterterpenes (4,5,10and11)were tested for their antibacterial,antifungal and cytotoxic properties,and six meroterpenoids (1-3,7-9) were evaluated for their antifungal activities.The antibacterial activity was evaluated against theEscherichiacoli,

Fig. 5 Experimental and calculated ECD curves of compound 6
Staphylococcusaureussubsp.Aureus,Salmonellaentericasubsp.Enterica andPseudomonasaeruginosa[29],but all tested compounds did not show any promising activity up to the highest tested concentration of 100 μM(Additional file 1: Table S7).Likewise,no cytotoxic activity against the human cancer cell lines (HL-60,A549,SMMC-7721,MDA-MB-231 and SW480) could be detected (at 40 μM) (Additional file 1: Table S8).The antifungal activity was evaluated against theEpidermophytonfloccosum,Trichophytonrubrum,Microsporum gypseumandCandidaalbicans.The compounds7,9,and10had good antifungal activity againstE.floccosumat a concentration of 100 μM,and compounds1,4,5and10exhibited moderate or weak antifungal activity againstM.gypseum(Table 3).
3 Conclusions
In conclusion,seventeen terpenoids,including six undescribed compounds,were identified from the endophytic fungusB.eleusines.Among these isolates,no compounds exhibited antibacterial and cytotoxic activities.However,the compounds7,9and10had significant antifungal activity againstE.floccosum.Our initial findings indicate that the endophytic fungusB.eleusinesis a plentiful supply of different and bioactive terpenoids.
3.1 General experimental procedures
A Bruker spectrometer (Bruker,Germany,model AM600) was used to obtain 1D and 2D spectra.A Q Exactive HF mass spectrometer (Thermo Fisher Scientific,USA) was used to collect HRESIMS date.Applied Circular dichroism spectrometer (Chirascan,New Haven,USA) was used to record CD spectra.Column chromatography (CC) was performed on silica gel (Qingdao Marine Chemical Ltd.,Qingdao,China),RP-18 gel(Fuji Silysia Chemical Ltd.,Japan),and Sephadex LH-20(Pharmacia Fine Chemical Co.,Ltd.,Sweden) [30].Semipreparative HPLC experiments were carried out on an Agilent 1260 HPLC with an Agilent Zorbax SB-C18column (5 μm,250 × 9.8 mm).Thin layer chromatography(GF 254) was used to monitor fractions,and spots were seen by heating silica gel plates coated with vanillin and 10% H2SO4in ethanol.
3.2 Culture and fermentation of fungal material
This fungus was identified by PCR amplification using fungal 18S universal primers ITS1 and ITS4.The sequenced 18S rDNA ITS sequences were subjected to homology sequence alignment analysis,which,using BLAST,showed 99% maximum similarity toBipolariseleusineswith the gene bank registration number(KY909768.1).Therefore,it was identified asB.eleusines,which was preserved in South-Central Minzu University,China.The strains were inoculated into PDA agar medium and incubated for 3-5 days at a constant temperature of 25 °C.The medium was then cut into several small pieces and placed into a solid wheat medium (50 g wheat and 50 mL water in 500 mL culture flasks,autoclaved at 120 °C for 30 min,for a total of 212 flasks) and fermented at 25 °C for 28 days.
3.3 Extraction and isolation
The wheat medium was pounded and extracted five times with EtOAc to yield 167 g of raw extract.The EtOAc extract was subjected to silica gel CC and eluted with a gradient of CH2Cl2-MeOH (100:0-0:100,ν/ν) to obtain six fractions (A-F).Fraction D (31.6 g) was subjected to C18 MPLC using MeOH-H2O (30:70-100:0,ν/ν),yielding ten subfractions (D1-D10).Fraction D5 (1.9 g) wasdivided by CC over silica gel and eluted in PE with a gradient of increasing acetone concentrations (30-100)to provide six fractions (D5-1-D5-6).Fraction D5-5(613.9 mg) was divided by using preparative HPLC (gradient elution with MeOH-H2O,18:82-40:60) to get11(3.2 mg,tR=12.7 min) and12(2.1 mg,tR=13.5 min).The Fraction D9 (903.5 mg) was separated into six subfractions D9-1-D9-6 by column chromatography (Sephadex LH-20,MeOH).Fraction D9-3 (113.6 mg) was separated by using preparative HPLC with a gradient of MeCN-H2O (50:50-60:40,ν/ν) to obtain8(4.3 mg,tR=20.5 min),9(1.2 mg,tR=24.6 min) and10(2.2 mg,tR=25.2 min).Fraction D9-4 (160.1 mg) was separated by using preparative HPLC with a gradient of MeCNH2O (40:60-60:40,ν/ν) to get7(3.5 mg,tR=12.1 min)and16(2.1 mg,tR=14.5 min).Fraction D9-6 (106.2 mg)was separated by Sephadex LH-20 (MeOH) to give four subfractions D9-6-1-D9-6-4 and then purified by usingpreparative HPLC with MeCN-H2O (50:50-60:40,ν/ν) to obtain2(12.2 mg,tR=32.6 min),14(4.5 mg,tR=34.2 min) and13(1.1 mg,tR=35.3 min).Fraction E(33.2 g) was separated by C18 MPLC using MeOH-H2O(10:90-100:0,ν/ν),yielding eight subfractions (E1-E8).The fraction E4 (3.4 g) was isolated by using Sephadex LH-20 and eluted with MeOH to yield five subdivisions E4-1-E4-5.Fraction E4-2 (532.6 mg) was divided by C18 MPLC with MeOH-H2O (60:40-75:25,ν/ν) to obtain3(5.2 mg,tR=18.2 min) and15(13.5 mg,tR=24.5 min).Fraction E4-5 (267.9 mg) was separated by using preparative HPLC with MeOH-H2O (65:35-80:20,ν/ν) to afford6(20.6 mg,tR=22.4 min) and4(15.7 mg,tR=26.8 min).Fraction E8 (7.4 g) was subjected to CC over silica gel with a gradient elution of PE-acetone (30:1-30:60,ν/ν)and then separated using a preparative C18 HPLC column with MeCN-H2O (45:55-60:40,ν/ν) to afford1(4.1 mg,tR=26.4 min),5(20.6 mg,tR=34.3 min) and17(6.2 mg,tR=36.4 min).

Table 3 Inhibitory effect of compounds on four strains of fungi
3.3.1 Bipolariterpene A (1)
Colorless solid;mp 247-252 ℃;UV (CH3OH)λmax(logε)=235 (3.61);IR (KBr): 3414,2970,2939,2874,1713,1655,1458,1431,1381,1342,1308,1246,1146,1092,1026,833 cm-1;[α]+361 (c0.5,CH3OH);1H NMR and13C NMR (CDCl3) data are shown in Table 1;HRESIMSm/z532.27271 [M+H]+(calcd for C29H42NO6S,532.27274).
3.3.2 Bipolariterpene B (2)
Yellow oil;UV (CH3OH)λmax(logε)=210 (2.49),225(2.49),275 (2.57);IR (KBr): 3248,2963,2936,1666,1632,1578,1493,1443,1377,1304,1269,1207,1138,1096,1053,1022,957 cm-1;[α]+315 (c0.5,CH3OH);1H NMR and13C NMR (CDCl3) data are shown in Table 1;HRESIMSm/z490.25650 [M+Na]+(calcd for C28H37NNaO5,490.25639).
3.3.3 Bipolariterpene C (3)
Purple solid;mp 259-264 ℃;UV (CH3OH)λmax(logε)=245 (2.81);IR (KBr): 3453,2959,2939,2874,2862,1666,1647,1628,1570,1516,1458,1381,1334,1227,1207,1142,1092,1022 cm -1;[α]+216 (c0.1,CH3OH);1H NMR and13C NMR (methanol-d4) data are shown in Table 1;HRESIMSm/z470.28995 [M+H]+(calcd for C28H40NO5,470.29010).
3.3.4 Bipolariterpene D (4)
Colorless powder;UV (CH3OH)λmax(logε)=205 (2.69),260 (2.55);IR (KBr): 3387,2974,2939,1740,1709,1655,1458,1373,1238,1038 cm-1;[α]+14 (c0.6,CH3OH);1H NMR and13C NMR (methanol-d4) data are shown in Table 2;HRESIMSm/z483.27190 [M+Na]+(calcd for C27H40NaO6,483.27171).
3.3.5 Bipolariterpene E (5)
Colorless powder;UV (CH3OH)λmax(logε)=265 (1.59);IR (KBr): 3375,2970,2936,1697,1651,1450,1408,1389,1312,1142,1030,941,856 cm-1;[α]+19 (c0.5,CH3OH);1H NMR and13C NMR (CDCl3) data are shown in Table 2;HRESIMS m/z 441.26097 [M+Na]+(calcd for C25H38NaO5,441.26115).
3.3.6 Bipolariterpene F (6)
Colorless powder;UV (CH3OH)λmax(logε)=210 (2.56);IR (KBr): 3314,2955,1659,1597,1450,1346,1177,1130,1042,1003,949,880 cm-1;[α]-2 (c0.5,CH3OH);1H NMR and13C NMR (methanol-d4) data are shown in Table 2;HRESIMSm/z237.18486 [M+H]+(calcd for C15H25O2,237.18491).
3.4 Quantum chemical calculations
3.4.1 13C NMR calculation
The NMR chemical shift calculations were carried out in Gaussian 16 using density functional theory (DFT) [31].Spartan’14 software was used to conduct the preliminary conformational distribution search.GaussView 6.0 was used to view conformational structures and modify computation input files for calculation.At the M062X/def-2svp level,all ground-state geometries were optimized,and the stable conformations obtained at that level were then used in magnetic shielding constants at the mpw1pw91/6-31+g(d,p) level.GaussView 6.0 was used to display the calculated chemical shift of each atom in each conformer,and the final chemical shift of each atom was calculated from the Boltzmann distribution of each conformer.The stereochemistry was assigned using the DP4+probability analysis of calculated and experimental chemical shifts.
3.4.2 ECD calculations
The absolute conformations of the compounds were determined using Gaussian 16 software.Briefly,the relative conformations of the compounds were first determined by ROESY spectroscopy,followed by a preliminary conformational analysis based on Spartan’14software.The obtained conformations were optimized at the M062X/def2svp level of density functional theory(TDDFT),and then ECD calculations were performed by M062X/def2svp.The ECD curves were generated by Origin software.
3.5 Antifungal assay
The endophytic fungus used in this assay wasBipolaris eleusines,and the samples to be tested were diluted in 96-well plates,and the fungal solution was added to each well.C.albicanswas nurtured at 37 °C for 24 h,and Filamentous fungi were nurtured at 25 °C for 5 days,and the absorbance value at 625 nm was measured by an enzyme marker.The experiment was set up with a medium blank control,a fungal control [32],and positive drug controls for amphotericin B and terbinafine hydrochloride.
3.6 Antibacterial assay
The samples were evaluated for their antibacterial activity againstEscherichiacoli,Staphylococcusaureussubsp.Aureus,Salmonellaentericasubsp.Enterica andPseudomonasaeruginosa.Dissolve the compounds to be tested to a concentration range of 100 μM and add it to a 96-well culture plate.Each well received a bacteria solution until the final concentration reached 5 × 105CFU/ml.It was then cultivated at 27 ℃ for 24 h,and the inhibition rate was determined by the microplate reader at OD 600 nm.The medium blank control was used in the experiment [12].Sodium penicillin G and ceftazidime were used as the positive control.
3.7 Cytotoxicity assay
Five human cancer cell lines including HL-60 (promyelocytic leukemia),A549 (lung epithelial cancer),SMMC-7721 (liver cancer),MDA-MB-231 (breast cancer),and SW480 (colon cancer) were used for cytotoxicity assays by MTS method [12].In a word,cells were cultured in DMEM media supplemented with 10% fetal bovine serum before being injected in 96-well culture plates and treated with different concentrations of compounds.After 24 h,each well received 20 μL of MTS,and the OD at 492 nm was determined using a microplate reader.
Supplementary Information
The online version contains supplementary material available at https:// doi.org/ 10.1007/ s13659-023-00407-x.
Additional file 1: Fig.S1.1H NMR spectrum of1.Fig.S2.13C NMR spectrum of1.Fig.S3.HSQC spectrum of1.Fig.S4.HMBC spectrum of1.Fig.S5.1H-1H COSY spectrum of1.Fig.S6.ROESY spectrum of1.Fig.S7.HRESIMS spectrum of1.Fig.S8.IR spectrum of1.Fig.S9.UV spectrum of1.Fig.S10.1H NMR spectrum of2.Fig.S11.13C NMR spectrum of2.Fig.S12.HSQC spectrum of2.Fig.S13.HMBC spectrum of2.Fig.S14.1H-1H COSY spectrum of2.Fig.S15.ROESY spectrum of2.Fig.S16.HRESIMS spectrum of2.Fig.S17.IR spectrum of2.Fig.S18.UV spectrum of2.Fig.S19.1H NMR spectrum of3.Fig.S20.13C NMR spectrum of3.Fig.S21.HSQC spectrum of3.Fig.S22.HMBC spectrum of3.Fig.S23.1H-1H COSY spectrum of3.Fig.S24.ROESY spectrum of3.Fig.S25.HRESIMS spectrum of3.Fig.S26.IR spectrum of3.Fig.S27.UV spectrum of3.Fig.S28.1H NMR spectrum of4.Fig.S29.13C NMR spectrum of4.Fig.S30.HSQC spectrum of4.Fig.S31HMBC spectrum of4.Fig.S32.1H-1H COSY spectrum of4.Fig.S33.ROESY spectrum of4.Fig.S34.HRESIMS spectrum of4.Fig.S35.IR spectrum of4.Fig.S36.UV spectrum of4.Fig.S37.1H NMR spectrum of5.Fig.S38.13C NMR spectrum of5.Fig.S39.HSQC spectrum of5.Fig.S40.HMBC spectrum of5.Fig.S41.1H-1H COSY spectrum of5.Fig.S42.ROESY spectrum of5.Fig.S43.HRESIMS spectrum of5.Fig.S44.IR spectrum of5.Fig.S45.UV spectrum of5.Fig.S46.1H NMR spectrum of6.Fig.S47.13C NMR spectrum of6.Fig.S48.HSQC spectrum of6.Fig.S49.HMBC spectrum of6.Fig.S50.1H-1H COSY spectrum of6.Fig.S51.ROESY spectrum of6.Fig.S52.HRESIMS spectrum of6.Fig.S53.IR spectrum of6.Fig.S54.UV spectrum of6.Fig.S55.1H NMR spectrum of7.Fig.S56.13C NMR spectrum of7.Fig.S57.1H NMR spectrum of8.Fig.S58.13C NMR spectrum of8.Fig.S59.1H NMR spectrum of9.Fig.S60.13C NMR spectrum of9.Fig.S61.1H NMR spectrum of10.Fig.S62.13C NMR spectrum of10.Fig.S63.1H NMR spectrum of11.Fig.S64.13C NMR spectrum of11.Fig.S65.1H NMR spectrum of12.Fig.S66.13C NMR spectrum of12.Fig.S67.1H NMR spectrum of13.Fig.S68.13C NMR spectrum of13.Fig.S69.1H NMR spectrum of14.Fig.S70.13C NMR spectrum of14.Fig.S71.1H NMR spectrum of15.Fig.S72.13C NMR spectrum of15.Fig.S73.1H NMR spectrum of16.Fig.S74.13C NMR spectrum of16.Fig.S75.1H NMR spectrum of17.Fig.S7613C NMR spectrum of17.Fig.S77.Correlations between calculated and experimental13C NMR chemical shifts of1Aand1B.Fig.S78.DP4+analysis results of1.Table S1.Energy analysis for conformers of1Aa~1Aeat mpw1pw91/6-31+g(d,p) level in the gas phase.Table S2.Cartesian coordinates for the low-energy optimized conformers of1Aat M062X/def2svp level.Fig.S79.Correlations between calculated and experimental13C NMR chemical shifts of2Aand2B.Fig.S80.DP4+analysis results of2.Table S3.Energy analysis for conformers of2Aa~2Aeat mpw1pw91/6-31+g(d,p) level in the gas phase.Table S4.Cartesian coordinates for the low-energy optimized conformers of2Aat M062X/def2svp level.Fig.S81.Correlations between calculated and experimental.13C NMR chemical shifts of4Aand4B.Fig.S82.DP4+analysis results of4Table S5.Energy analysis for conformers of4Aa~4Aeat mpw1pw91/6-31+g(d,p) level in the gas phaseTable S6.Cartesian coordinates for the low-energy optimized conformers of4Aat M062X/def2svp level.Table S7.Inhibitory effect of compounds on four strains of bacteria.Table S8.Inhibitory effect of compounds on five strains of Cytotoxicity.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (32000011,82204239),Hubei Provincial Natural Science Foundation of China (2022CFB462),the Fund of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2022-KF03),the Fundamental Research Funds for the Central Universities,South-Central MinZu University(CZY23024).The authors thank the Bioactivity Screening Center,Kunming Institute of Botany,Chinese Academy of Sciences,for screening the bioactivity of the compounds.
Author contributions
H.-L.A.isolated and provided the fungus.Y.-Z.F.contributed to the extraction,isolation,and identification of the samples.C.T.was responsible for the evaluation of cytotoxic activity.S.-Y.T.,Q.L.and F.X.contributed to the extraction and isolation of the samples.Y.-Z.F.and B.-B.S.performed chemical calculation and wrote the paper.H.-L.A.and J.-K.L.designed experiments.All authors reviewed the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32000011,82204239),Hubei Provincial Natural Science Foundation of China (2022CFB462),the Fund of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2022-KF03),the Fundamental Research Funds for the Central Universities,South-Central MinZu University(CZY23024).
Availability of data and materials
All data generated and analyzed during this study are included in this published article and its Additional file.
Declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author details
1School of Pharmaceutical Sciences,South-Central MinZu University,Wuhan 430074,People’s Republic of China.
Received:19 July 2023
Accepted:8 October 2023

杂志排行
Natural Products and Bioprospecting的其它文章
- Quinones from Cordia species from 1972 to 2023: isolation,structural diversity and pharmacological activities
- Ginsenoside compound-K attenuates OVX-induced osteoporosis via the suppression of RANKL-induced osteoclastogenesis and oxidative stress
- The alkynyl-containing compounds from mushrooms and their biological activities
- A recent update on development,synthesis methods,properties and application of natural products derived carbon dots
- Natural product rhynchophylline prevents stress-induced hair graying by preserving melanocyte stem cells via the β2 adrenergic pathway suppression
- Kaemtakols A-D,highly oxidized pimarane diterpenoids with potent anti-inflammatory activity from Kaempferia takensis
