High-resolution genetic mapping and identification of candidate genes for the wheat stem rust resistance gene Sr8155B1
2023-12-25JianWangHongyuLiTaoShenShikaiLyuShamsurRehmanHongnaLiGuipingWangBinyangXuQingWangWanyiHuKairongLiShengshengBaiJianMaHaitaoYuMatthewRouseShishengChena
Jian Wang,Hongyu Li,Tao Shen,Shikai Lyu,Shams ur Rehman,Hongna Li,Guiping Wang,Binyang Xu, Qing Wang, Wanyi Hu, Kairong Li, Shengsheng Bai, Jian Ma, Haitao Yu,Matthew N.Rouse*, Shisheng Chena,,*
a School of Advanced Agriculture Sciences, Peking University, Beijing 100871, China
b National Key Laboratory of Wheat Improvement,Peking University Institute of Advanced Agricultural Sciences,Shandong Laboratory of Advanced Agricultural Sciences in Weifang,Weifang 261325, Shandong, China
c Triticeae Research Institute, Sichuan Agricultural University, Chengdu 611130, Sichuan, China
d Wheat Research Institute, Weifang Academy of Agricultural Sciences, Weifang 261071, Shandong, China
e US Department of Agriculture-Agricultural Research Service, Cereal Disease Laboratory and Department of Plant Pathology, University of Minnesota, St.Paul, MN 55108, USA
Keywords:Durum wheat Stem rust Resistance gene Sr8155B1 CC-NBS-LRR
ABSTRACT Stem rust,caused by Puccinia graminis f.sp.tritici(Pgt),threatens global wheat production.Development of cultivars with increased resistance to stem rust by identification, mapping, and deployment of resistance genes is the best strategy for controlling the disease.In this study, we performed fine mapping and characterization of the all-stage stem rust resistance (Sr) gene Sr8155B1 from the durum wheat line 8155-B1.In seedling tests of biparental populations,Sr8155B1 was effective against six Chinese Pgt races tested.In a segregating population of 5060 gametes, Sr8155B1 was mapped to a 0.06-cM region flanked by markers Pku2772 and Pku43365, corresponding to 1.5- and 2.7-Mb regions in the Svevo and Chinese Spring reference genomes.Both regions include several typical nucleotide-binding leucine-rich repeat(NLR)and protein kinase genes that represent candidate genes.Among them,three NLR genes and three receptor-like protein kinases were highly polymorphic between the parental lines and their transcripts were upregulated in the homozygous resistant line TdR2 relative to its susceptible sister line TdS4.Four markers(Pku2772,Pku43365,Pku2950,and Pku3721)developed in this study,together with seedling resistance responses,correctly predicted Sr8155B1 absence or presence in 78 tetraploid wheat genotypes tested.The presence of Sr8155B1 in tetraploid wheat accessions CItr 14916,PI 197492,and PI 197493 was confirmed by mapping in three F2 populations.The genetic map and linked markers developed in this study may accelerate the deployment of Sr8155B1-mediated resistance in wheat breeding programs.
1.Introduction
Stem (black) rust is a devastating foliar disease caused by the fungal pathogen Puccinia graminis f.sp.tritici (Pgt) that threatens the productivity of bread wheat, Triticum aestivum L.(2n=6x=42,AABBDD)and durum wheat,Triticum turgidum subsp.durum(Desf.)Husn.(2n=4x=28,AABB).In the 19th and early 20th centuries, severe stem rust epidemics in North America, Europe,and other wheat-growing regions incurred yield reductions and economic losses [1,2].In recent decades, stem rust has been controlled mainly by the deployment of resistance genes, elimination of the pathogen’s alternative host, barberry (Berberis vulgaris L.),and monitoring of pathogen populations for potentially aggressive new races [3,4].
Stem rust has again become a threat with the emergence of the Pgt race TTKSK (also called Ug99) in East Africa in 1998 [5].Ug99 was the first known Pgt race that could overcome resistance conferred by the resistance gene Sr31 [5], and subsequent variants of Ug99 overcame additional Sr genes, including Sr9h, Sr24, Sr36,and SrTmp [6–9].To date, there are 13 known variants belonging to the Ug99 race group, which has migrated across eastern and southern Africa [3,10,11], into the Middle East [12,13], and has spread to at least 13 countries [1].Given that about 90 % of the world’s wheat cultivars are susceptible to Ug99 or its variants,the Ug99 race group threatens world wheat production [14].
In recent years, other virulent Pgt pathotypes (TKTTF, TRTTF,and TTRTF)unrelated to the Ug99 race group but exhibiting broad virulence have been identified in stem rust outbreaks.The TKTTF race caused severe epidemics in Ethiopia in 2013 and 2014 owing to its virulence to the resistance gene SrTmp in the wheat cultivar Digalu [15].This race has been found in more than ten countries,including Iran, Turkey, Lebanon, Sweden, Denmark, and Germany[16,17].Race TRTTF is virulent to at least four Sr genes that are effective against race TTKSK: Sr13, Sr36, SrTmp, and Sr1RSAmigo[3].Race TTRTF was first found in Georgia in 2014 [18], later caused a stem rust epidemic on durum wheat in Sicily in 2016 [19], and was virulent to 23 Sr genes [20].Given the evolution and spread of new Pgt races, there is an urgent need to identify new Sr genes and to diversify the combinations of deployed genes in wheat breeding programs.
To date, over 60 Sr genes (Sr1–Sr63) have been assigned official designations in wheat and its wild relatives[21].Tetraploid wheat(T.turgidum ssp.), which is part of the primary gene pool of common wheat, has contributed several Sr genes, including Sr2, Sr9d/Sr9e/Sr9g/SrKN, Sr11, Sr12, Sr13a/Sr13b, Sr14, Sr17, and Sr8155B1[14,22–24].However, among the Sr genes derived from tetraploid wheat, only two genes, Sr9 and Sr13, have been cloned [24,25],owing mainly to the size and complexity of wheat genomes.
The all-stage resistance gene Sr8155B1,discovered in the durum wheat line 8155-B1, is effective against several Pgt races tested,including TTKTT, TTTSK, TTKSF+, TRTTF, TTKST, and TMLKC, but is susceptible to Pgt races TTKSK and JRCQC [22].This gene was previously mapped within a 2.5-cM interval on the very distal region of the short arm of chromosome 6A closely linked to markers KASP_6AS_IWB61585, KASP_6AS_IWB10558, and KASP_6AS_IWB1550[22].When introgressed into bread wheat, Sr8155B1 conferred strong resistance against Pgt[22].The objectives of this study were to 1)test whether Sr8155B1 confers resistance to Chinese Pgt races,2)generate a high-density genetic map of Sr8155B1,and 3)identify candidate genes for Sr8155B1.
2.Materials and methods
2.1.Plant materials and mapping populations
The durum wheat line 8155-B1 was crossed with Rusty,a tetraploid genetic stock susceptible to almost all Pgt races[26].For initial mapping of Sr8155B1,a subset of 294 F2plants from the 8155-B1×Rusty cross was inoculated with Pgt race 34C3RTGQM(isolate 20IAL32).After the initial mapping,12 F2plants(plants 3,5,16,26,62,69,74,106,114,124,134,and 220)that were heterozygous for the Sr8155B1 region were selected using flanking markers and a second segregating population of 2236 F3plants was generated.These F3plants were screened with flanking markers to identify plants with recombination events and the progeny of the recombinants were challenged with Pgt race 34C3RTGQM.The markers closely linked to Sr8155B1 were employed in marker-assisted selection in a collection of 78 T.turgidum ssp.dicoccon accessions.Five of these accessions (CItr 14916, PI 197486, PI 197492, PI 197493,and PI 387683)were crossed separately with Rusty to generate five F2segregating populations.
2.2.Stem rust assays
Infection types (ITs) of the parental lines 8155-B1 and Rusty against Pgt races TMLKC (isolate 72-41-Sp2), TTKST (isolate 06KEN19v3), TTKSK (04KEN156/04), TTKTT (14KEN58-1), TTTSK(07KEN24-4), TTTSK+ (09ZIM01-2), TRTTF (06YEM34-1), and JRCQC (08ETH03-1) have been previously reported [22].In this study, stem rust seedling assays were performed at the Peking University Institute of Advanced Agricultural Sciences using the Chinese Pgt races as described previously [27].The parental lines and selected homozygous resistant or susceptible F2:3families were inoculated with six Chinese Pgt races: 34C3RTGQM (isolate 20IAL32), 34MKGQM(20IAL06),34MTGSM(20GSA1),21C3CTTTM(20GH13), RTGRM (20PKU3), and 34C3RKGQM (21PK9).The virulence/avirulence formulae of these races are presented in Table S1.Plants were grown in growth chambers at 22 °C during the day and 20 °C at night under a 16:8h light/dark photoperiod.ITs were scored using a 0–4 scale as described previously [27].
2.3.Marker development
For development of DNA markers in the 8155-B1 × Rusty population, whole-genome resequencing of both parents was performed.The raw sequencing data was deposited at the National Genomics Data Center under BioProject accession number PRJCA017761.Genomic library preparation and sequencing were performed by Novogene Bioinformatics Technology Co., Ltd.(Beijing, China).The 8155-B1 and Rusty sequences of the genes in the candidate region on chromosome arm 6AS were aligned, and polymorphic sites were identified.Sequence alignment, quality control, and variant calling were performed as previously described [28].Genome-specific primer pairs were designed with Primer 3 (https://bioinfo.ut.ee/primer3-0.4.0/primer3/) to amplify regions carrying target polymorphisms.Genotypes of the parents and segregating populations were acquired using either the Kompetitive allele-specific polymerase chain reaction(KASP)or cleaved amplified polymorphic sequence (CAPS) markers.KASP assays were performed according to the manufacturer’s protocol (LGC Genomics; https://www.lgcgroup.com), and the fluorescent signal was detected using a BMG FLUOstar Omega plate reader (BMG LABTECH GmbH, Offenburg, Germany).Amplification and enzyme cleavage reactions of the CAPS markers were performed according to standard procedures for the corresponding restriction endonucleases (New England Biolabs, Hitchin, UK).
2.4.RNA-seq and qRT-PCR
From the 8155-B1 × Rusty population, a pair of F4sister lines homozygous for the presence (TdR2) or absence (TdS4) of Sr8155B1 were developed using the identified markers.Seedlings of TdR2 and TdS4 were inoculated with Pgt race 34C3RTGQM and leaf samples from multiple plants were collected at 6 d postinoculation (dpi).Pgt-inoculated leaves were used for RNA-seq because we did not know whether or not the Sr8155B1 gene is induced by the pathogen.Three biological replicates were used for each genotype.RNA-seq library preparation and sequencing were performed by Novogene.The raw sequencing data is available under the BioProject Accession Number PRJCA017761.Differentially expressed genes (DEGs) were identified with edgeR [29]using parameters P-value < 0.05, |log2fold change| > 1, and false discovery rate (FDR) < 0.05.DEGs within the Sr8155B1 candidate region of chromosome 6A were identified,and a heatmap was generated with the pheatmap R package [30].The same RNA samples were used to quantify the transcript levels of candidate genes located in the Sr8155B1 candidate region.qRT-PCR was performed on an Applied Biosystems QuantStudio 5 Real-Time PCR System using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific,Waltham, MA, USA).Transcript levels were expressed as fold-ACTIN levels using the 2-ΔCT method as described previously [24].
2.5.Statistical analyses
The published reference genomes of the hexaploid wheat Chinese Spring (CS) [31] and tetraploid wheat Svevo [32] were used to assist in marker development.The Wheat Expression Browser(expVIP, https://www.wheat-expression.com/) was used to detect the expressions of candidate genes[33].Linkage maps were drawn using MapChart version 2.2 software [34].The diagnostic marker Sr13F/R for Sr13 [24] and four pairs of gene-specific primers(pku4861F7R7, TRI2B223210F5R5, TRI2B223210F7R7, and TRI2B223210F15R15) for Sr9e [25] were reported previously.
3.Results
3.1.Seedlings of 8155-B1 showed high resistance to all Chinese Pgt races
We confirmed the absence of Sr13 and Sr9e in the resistant parent 8155-B1 using published diagnostic or gene-specific markers for Sr13 and Sr9e.Seedlings of the parental lines 8155-B1 and Rusty were challenged with six Pgt races collected in China (Table S1).8155-B1 showed strong levels of resistance (ITs = 0 or 0;) to all Pgt races tested, whereas Rusty showed susceptible ITs ranging from 3+to 4(Fig.1).The F1plants derived from the 8155-B1×Ru sty cross displayed a clear resistance phenotype in response to the Pgt race 34C3RTGQM.In a subset of 294 F2plants from the 8155-B1 × Rusty cross inoculated with race 34C3RTGQM, plants with ITs ranging from 0 to 1;were classified as resistant and those with ITs between 3+and 4 were classified as susceptible.In this population,232 plants were resistant and 62 plants were susceptible,corresponding to a segregation of 3:1(resistant:susceptible)expected for a single dominant genetic locus (χ2= 2.40, P = 0.12).
Using the markers KASP_6AS_IWB61585 and KASP_6AS_IWB1550 flanking Sr8155B1 [22], 10 F2:3families homozygous for the Sr8155B1 allele and 10 families homozygous for the susceptible Rusty allele were selected from the 8155-B1 × Rusty cross.These families were inoculated with four Chinese Pgt races(34C3RTGQM,34MKGQM, 34MTGSM, and 21C3CTTTM) and maintained in four separate growth chambers.All 10 families homozygous for the 8155-B1 allele were resistant to all four of the tested Pgt races,whereas all 10 families homozygous for the Rusty allele were susceptible to these races(Fig.S1),suggesting that 8155-B1 resistance to these races is linked to the Sr8155B1 region.
3.2.High-resolution genetic map of Sr8155B1
To develop molecular markers in the Sr8155B1 mapping region,we first performed whole-genome resequencing for 8155-B1 and Rusty to identify polymorphisms between them.Respectively 1041 million and 1124 million PE150 reads were generated for 8155-B1 and Rusty.Alignment of the cleaned sequences of 8155-B1 and Rusty revealed 10.5 million polymorphic sites distributed across all chromosomes.On chromosome 6A, 791,121 polymorphisms were detected between the two parents.
Based on the polymorphisms on chromosome arm 6AS,12 DNA markers (Table 1) were developed in the Sr8155B1 candidate region and a preliminary linkage map using the 294 phenotyped F2plants was generated (Fig.2A).Based on the linkage results,the 34C3RTGQM resistance gene was mapped to a 0.51-cM region between markers Pku2378 and Pku49163 (2.38–4.91 Mb in Svevo,Fig.2A).
To better define the position of Sr8155B1, we screened an additional 2,236 F2plants from 12 selected segregating F3families with the new flanking markers Pku2378 and Pku49163.This screening yielded 13 plants carrying recombination events.The genetic distance between Pku2378 and Pku49163 was re-estimated to be 0.32 cM based on 13 plants with recombination events identified in this screening and three plants found in the initial screening of the 294 plants.Inoculations with race 34C3RTGQM were performed on the progeny of plants carrying informative recombination events (~25 plants per family).Using these critical recombinants and 10 new markers developed in this region(Table 1), Sr8155B1 was further confined to a 0.06-cM interval flanked by markers Pku2772 and Pku43365 and was completely linked to markers Pku2950, Pku3107, Pku3590, Pku3721,Pku40006, and Pku40013 (Fig.2B).PCR amplifications with three KASP markers(Pku2772,Pku3107,and Pku3590)and the CAPS marker Pku43365 digested with StyI are shown in Fig.S2.
3.3.Candidate genes for Sr8155B1 in the colinear regions of tetraploid and hexaploid wheat genomes
The 0.06-cM candidate interval between marker loci Pku2772 and Pku43365 defines a 1.57-Mb genomic region in the reference genome of Svevo (2.77–4.34 Mb, Fig.2C; Table S2) and a 2.65-Mb region in CS (3.85–6.50 Mb, Table S3).The candidate regions contain 32 annotated high-confidence genes in Svevo (TRITD6Av1G001370–TRITD6Av1G001990) and 41 in CS (TraesCS6A01G009200–TraesCS6A01G013200).These genes include respectively five and seven typical NBS-LRR (NLR) genes in Svevo and CS (Tables S2, S3).This finding is particularly noteworthy because NLRs are the most prevalent class of genes associated with plant disease resistance [23–25,35–37].
DEGs between homozygous resistant (F4line TdR2) and homozygous susceptible(F4sister line TdS4)plants were identified from RNA-seq for Pgt-inoculated RNA samples of TdR2 and TdS4(Fig.3).Among the 32 candidate genes in the target interval in Svevo,13 were differentially expressed(FDR<0.05,P-value<0.05,and |log2fold change| > 1) between the Pgt-inoculated susceptible and resistant plants (Fig.3; Table S4).Twelve DEGs were significantly upregulated in plants carrying the resistant Sr8155B1 allele relative to those carrying the susceptible allele, whereas only one DEG was downregulated between the two groups(Table S4).These upregulated DEGs included three annotated NLR genes (TRITD6Av1G001430, TRITD6Av1G001520, and TRITD6Av1G001530) and three receptor-like protein kinases (TRITD6Av1G001460, TRITD6Av1G001470, and TRITD6Av1G001990), which represent good candidate genes.The transcript levels of these six candidate genes were determined by qRT-PCR(Table S5).The transcript levels of all six candidate genes were significantly higher (P < 0.001) in TdR2 than in TdS4 (Fig.S3), supporting the findings from RNA-seq analysis.
Using the whole-genome resequencing reads from the 8155-B1 and Rusty parental lines, we assembled the reads and obtained genomic contigs containing the candidate genes.Comparison of the obtained genomic contigs of 8155-B1 and Rusty revealed that four candidate genes (TRITD6Av1G001460, TRITD6Av1G001470,TRITD6Av1G001520,and TRITD6Av1G001530)were deleted in Rusty(Fig.S4).The other two candidate genes, TRITD6Av1G001430 and TRITD6Av1G001990, showed very low similarity between the two parents (Fig.S5).
3.4.Validation of Sr8155B1-linked markers in uncharacterized tetraploid wheat genotypes

Fig.1.Susceptibility/resistance responses of 8155-B1 and Rusty to Pgt races(A)34C3RTGQM,(B)34MKGQM,(C)34MTGSM,(D)21C3CTTTM,(E)RTJRM,and(F)34C3RKGQM.Plants were grown in growth chambers at 22 °C during the day and 20 °C at night under a 16:8h light/dark photoperiod.RP, resistant parent; SP, susceptible parent.
To identify PCR markers able to distinguish genotypes with and without Sr8155B1, we evaluated a collection of 78 accessions of T.turgidum ssp.dicoccon with two flanking markers Pku2772 and Pku43365, and two completely linked markers Pku2950 and Pku3721.The T.turgidum ssp.dicoccon lines were challenged with Chinese Pgt race 34MKGQM and their infection responses were listed in Table S6.Eight T.dicoccon accessions,including CItr 14916,PI 154582,PI 197481,PI 197486,PI 197492,PI 197493,PI 221400,and PI 387683,showed haplotypes identical to 8155-B1(Table S6).Of these, six (CItr 14916, PI 197486, PI 197492, PI 197493, PI 221400, and PI 387683) displayed similar resistance responses(ITs = 0 or 0;) to Pgt race 34MKGQM, suggesting that these lines might carry Sr8155B1.By contrast, the other two accessions (PI 154582 and PI 197481) had the same haplotype as 8155-B1 but lower levels of resistance than 8155-B1, suggesting the absence of Sr8155B1.
Another five pairs of genome-specific primers were developed in the Sr8155B1 candidate region (Table S7) to evaluate the six selected accessions.Sequencing of the PCR products revealed that all accessions but PI 221400 were identical to those of 8155-B1(Fig.S6).
3.5.Confirmation of the presence of Sr8155B1 in selected T.dicoccon accessions
Five F2segregating populations were generated from crosses between the susceptible line Rusty and the selected 34MKGQMresistant T.dicoccon accessions CItr 14916, PI 197486, PI 197492,PI 197493, and PI 387683.Approximately 70 F2plants from each population were inoculated with Pgt race 34C3RTGQM.In a subset of 72 F2plants from the CItr 14916 × Rusty cross inoculated with race 34C3RTGQM,55 plants were resistant and 17 plants were susceptible(Fig.4A),fitting the 3:1(resistant:susceptible)segregation ratio expected for a single dominant gene (χ2= 0.07, P = 0.79;Table S8).In the populations of PI 197492 × Rusty and PI 197493 × Rusty, the segregation of phenotypes resistant and susceptible to race 34C3RTGQM also fitted that of a single dominant gene (Fig.4B, C; Table S8).Genotyping these F2plants with markers Pku2772 and Pku43365 flanking Sr8155B1 revealed that the 34C3RTGQM resistance phenotypes in these three mapping populations were completely linked to the Sr8155B1 locus.These results suggested that the observed resistance to 34C3RTGQM in the T.dicoccon accessions CItr 14916, PI 197492, and PI 197493 was conferred by Sr8155B1.
In the PI 197486×Rusty and PI 387683×Rusty mapping populations, segregations of resistance to race 34C3RTGQM fitted a two-gene model (Table S8).Genotyping with Sr8155B1-flanking markers Pku2772 and Pku43365 showed that all susceptible plants(ITs = 3 to 4) in both populations carried the susceptible Sr8155B1 allele,indicating the presence of Sr8155B1 and another unknown Sr gene.However, given the small size of the mapping populations,further studies will be needed to confirm the presence of Sr8155B1 in PI 197486 and PI 387683.
3.6.Comparison of Sr8155B1 and Sr8 located on chromosome arm 6AS
The only formally named wheat stem rust resistance gene on chromosome arm 6AS is Sr8 [38,39].Sr8 was located 4.6 ± 1.0 cM proximal to the simple sequence repeat (SSR) marker gwm334(7.3 Mb in Svevo RefSeq v1.0) [40], indicating that Sr8155B1(2.77–4.34 Mb in Svevo) and Sr8 mapped to the same chromosomal region.The Sr8155B1-linked markers Pku2772, Pku43365,Pku2950, and Pku3721 were genotyped in the Sr8a containing line ISr8a-Ra(Table S6).8155-B1 and ISr8a-Ra showed identical haplotypes,indicating that these markers are not sufficient to differentiate these genes or alleles.
Seedlings of ISr8a-Ra,8155-B1,and Rusty were challenged with six Chinese Pgt races(Table 2).ISr8a-Ra was susceptible(ITs=3 to 4)against all Pgt races tested,while 8155-B1 showed strong resistance(ITs= 0 or 0;)against the same races(Table 2).The differing resistance profiles of Sr8155B1 and Sr8a indicated that they are different genes or alleles.
4.Discussion
4.1.Inheritance of stem rust resistance in the durum wheat line 8155-B1
Nirmala et al.[22]reported that nine of 11(82%)durum wheat cultivars in the Great Plains of the USA carried the Sr8155B1 gene based on the haplotype of linked markers along with similar seedling resistance responses and resistance profiles.Aoun et al.[41]found that Sr8155B1 was widely distributed in the Midwest region of the USA with ~79%of durum wheat cultivars and breeding lines carrying this gene.In contrast,Sr8155B1 was present in only three T.dicoccon accessions (3.8%) in our study.The frequency of the Sr8155B1 gene is much lower in T.dicoccon than in T.durum, suggesting that Sr8155B1 may have undergone strong positive selection in modern durum wheat cultivars in the USA.
Segregation for seedling resistance in the F2population of the 8155-B1 ×Rusty cross showed that a single dominant gene confers resistance to Pgt race 34C3RTGQM.Dominant inheritancewas also attributed to the resistance phenotype in the F1plants.However, segregation of resistance based on previous genetic analyses of BC1F2and F2populations revealed Sr8155B1 to be a recessive gene [22].These contradictory or inconclusive results may be explained by dominance reversal.Dominance reversals have been reported for several rust resistance genes in wheat,such as Yr6 [42] and Sr6 [43].Although it has been hypothesized[43–47]that temperature,light,and different Pgt isolates account for dominance reversal, a definitive cause has yet to be confirmed.

Table 1 Primers used for fine mapping of Sr8155B1.
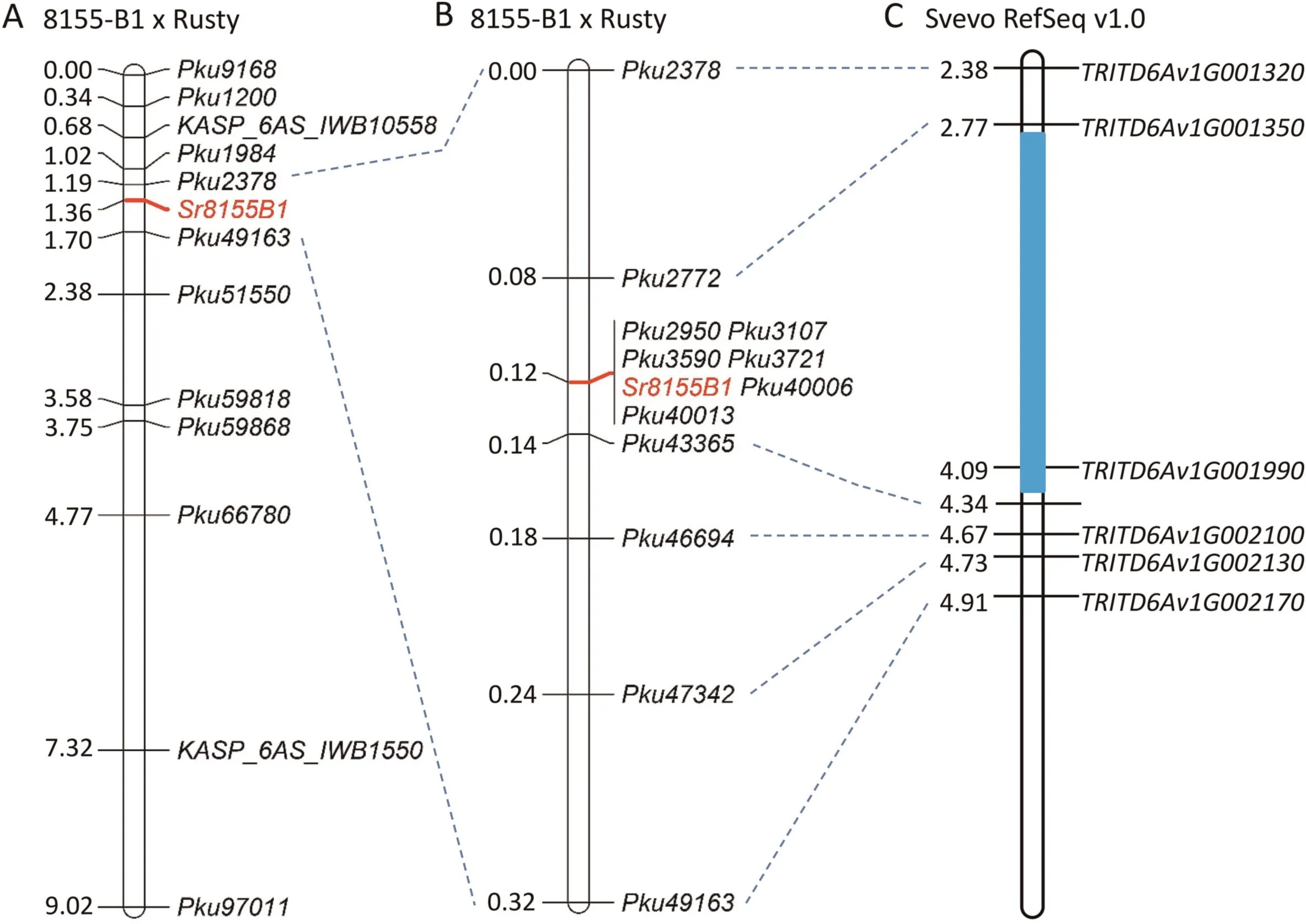
Fig.2.Fine mapping of the stem rust resistance gene Sr8155B1.(A)Genetic map based on 294 F2 plants and 12 DNA markers;(B)High-density genetic map based on 2,530 F2 plants and 10 new markers; (C) Colinear region in the sequenced Svevo reference genome.The values to the left of the DNA markers are the genetic distances (cM;centimorgans), while the values to the left of the genes are physical locations (Mb; Svevo RefSeq v1.0).
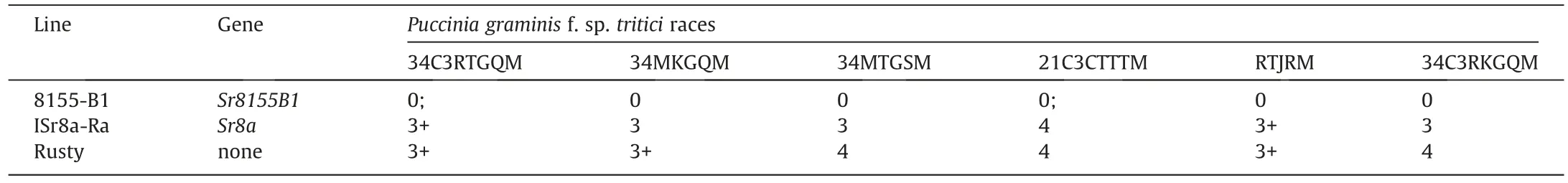
Table 2 The resistance profiles of Sr8155B1 and Sr8a against six Chinese Pgt races.
4.2.High-density mapping and identification of candidate genes for Sr8155B1
By leveraging the published reference genomes of tetraploid and hexaploid wheat,we narrowed the region of the Sr8155B1 candidate gene to a 1.57-Mb region in tetraploid wheat Svevo and a 2.65-Mb region in hexaploid wheat CS that includes a cluster of typical NLR genes.Clustering of NLR genes promotes the generation of novel variants by recombination, deletion/duplication, and conversion events, increasing the genetic variation of NLR genes[35,48].In the Sr8155B1 candidate region, several NLR genes appeared to be absent from the susceptible parent Rusty(Fig.S4),and NLR genes varied in both copy number and structure between hexaploid wheat CS and tetraploid wheat Svevo (Tables S2, S3).As in the Sr8155B1 region, rearrangements, deletions, and duplications of NLR genes have been reported for other Sr genes,such as SrKN [23], Sr21 [35], and Sr13 [24].
Many of the cloned disease resistance genes in wheat and its wild relatives encode intracellular NLR proteins [24,25,35–37,49,50],which recognize specific pathogen effectors and activate effector-triggered immunity.We hypothesize that one of the NLR genes in the target region is a candidate for the race-specific gene Sr8155B1.In addition to these NLR genes, we identified several receptor-like protein kinases (Table S4), which were completely linked to Sr8155B1 and differentially expressed in the resistant and susceptible lines (Fig.3).Receptor-like protein kinases could be candidates, as this type of protein has also been implicated in disease resistance in wheat and other plant species [51–57].All cloned race-specific Sr genes encode NLR proteins except for Sr60, which encodes a protein kinase [52].Sr8155B1 may encode a protein kinase,given that protein structure is insufficient to predict race specificity [52].
4.3.Relationship between Sr8155B1 and other Sr genes on chromosome 6A
Chromosome 6A harbors several formally named Sr genes,including Sr8, Sr13, Sr26, and Sr52 [38,58,59].Of these, Sr13, Sr26,and Sr52 were located on the long arm of chromosome 6A and conferred seedling resistance to Pgt race TTKSK [24,58–60].Sr26 and Sr52 were introgressed into wheat from the wild species Thinopyrum ponticum(Podp.)Barkworth&D.R.Dewey and Dasypyrum villosum (L.) Candargy, respectively [61–63].The absence of Sr13 in the resistant parent 8155-B1 was confirmed using a published diagnostic marker for Sr13 [24].These results demonstrated that Sr8155B1 is different from the Sr13, Sr26, and Sr52 genes.
The Sr8 alleles,including Sr8a and Sr8b,were mapped to the distal region of chromosome arm 6AS [38,39] but showed a very different resistance profile than Sr8155B1 [22] (Table 2), indicating that they are different genes or alleles.The lack of markers linked to both Sr8 and Sr8155B1 and the absence of high-density genetic maps of Sr8 prevented establishing the mapping relationship between these two genes.A more precise genetic map of Sr8 that shares PCR markers with the Sr8155B1 map is needed to determine whether Sr8155B1 and Sr8 are different alleles of the same gene or different genes.Alternatively, allelism tests of Sr8155B1 and Sr8 could establish these relationships.
Several temporarily designated Sr genes have also been mapped on chromosome arm 6AS: Sr_TRTTF, QSr.sun-6AS, and QSr.cdl-6A[39,60,64].Among them, Sr_TRTTF and QSr.sun-6AS were mapped in hexaploid but not in tetraploid wheat, and were concluded to be Sr8a [39,64].QSr.cdl-6A, which confers adult plant resistance,was mapped in the hexaploid wheat line CI 14275 and explained 6.4%to 12.6%of phenotypic variation in various field environments[60].QSr.cdl-6A was located between 51.4 and 52.0 Mb (CS RefSeq v1.0) [60], which is in a different location than Sr8155B1 (3.85–6.50 Mb).In addition,the susceptibility of CI 14275(the donor line of QSr.cdl-6A)and the derived RIL population to the Pgt race TRTTF rules out the possibility of QSr.cdl-6A being Sr8155B1 [60].
4.4.Conclusions and practical implications
Sr8155B1 confers strong levels of resistance against multiple Pgt races from China (the current study), North America, and Africa[22].The strong resistance conferred by Sr8155B1 makes it a potentially valuable genetic resource in wheat breeding.However,because Sr8155B1 is not effective against several Pgt races, including the original Ug99 race TTKSK [22], it would need to be deployed in combination with other Sr genes.Some potentially valuable Sr genes that could expand the resistance spectrum include Sr13 [24], Sr35 [37], Sr22a/Sr22b [36,65], and Sr33 [66],which are all resistant to the Pgt race TTKSK.
Direct hybridization without special cytogenetic manipulation can be used to transfer genes from T.durum into hexaploid bread wheat, given that they share common A and B genomes [23].Examples of successful mapping and transfer of rust resistance genes from durum to bread wheat include the stem rust resistance genes Sr13 [62]and SrKN [23];the stripe rust resistance genes Yr5[67], Yr64, and Yr65 [68]; and the leaf rust resistance genes Lr23[69], Lr61 [70], and Lr79 [71].To accelerate the deployment of Sr8155B1 in common wheat breeding programs, we initiated the introgression of this gene into several Chinese bread wheat varieties.We used the closely linked markers for Sr8155B1 developed in this study(Table 1)to validate the presence of the 8155-B1 segment during backcrossing.
In conclusion, the high-density map of Sr8155B1 together with the tightly linked PCR markers identified in this study will facilitate map-based cloning of this gene and accelerate its deployment and pyramiding with other Sr genes.
CRediT authorship contribution statement
Jian Wang:Data curation, Investigation, Formal analysis, Writing-Original draft preparation.Hongyu Li:Data curation,Investigation, Formal analysis, Writing - Original draft preparation.Tao Shen:Data curation,Investigation,Formal analysis,Writing-Original draft preparation,Validation.Shikai Lyu:Validation,Writing-Original draft preparation.Shams ur Rehman:Validation,Writing-Original draft preparation.Hongna Li:Software,Formal analysis,Visualization.Guiping Wang:Software,Formal analysis,Visualization.Binyang Xu:Software, Formal analysis, Visualization.Qing Wang:Software, Formal analysis, Visualization.Wanyi Hu:Software, Formal analysis, Visualization.Kairong Li:Software, Formal analysis,Visualization.Shengsheng Bai:Software,Formal analysis,Visualization.Jian Ma:Resources.Haitao Yu:Resources.Matthew N.Rouse:Project administration, Methodology, Funding acquisition,Writing-reviewing&editing.Shisheng Chen:Project administration,Methodology,Funding acquisition,Writing-reviewing&editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2022YFD1201300), the Key R&D Program of Shandong Province (ZR202211070163), the Provincial Natural Science Foundation of Shandong (ZR2021ZD30,ZR2021MC056),and the Young Taishan Scholars Program of Shandong Province.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.09.006.
杂志排行
The Crop Journal的其它文章
- OsSPL10 controls trichome development by interacting with OsWOX3B at both transcription and protein levels in rice (Oryza sativa L.)
- Ectopic expression of OsNF-YA8, an endosperm-specific nuclear factor Y transcription-factor gene, causes vegetative and reproductive development defects in rice
- Mechanisms of autophagy function and regulation in plant growth,development, and response to abiotic stress
- ZmDRR206 functions in maintaining cell wall integrity during maize seedling growth and defense response to external stresses
- The plasmodesmata-associated β-1,3-glucanase gene GhPdBG regulates fiber development in cotton
- The MabHLH11 transcription factor interacting with MaMYB4 acts additively in increasing plant scopolin biosynthesis
