GmTOC1b negatively regulates resistance to Soybean mosaic virus
2023-12-25YuhngZhngHipingDuTintinZhoChunmeiLioTuFengJunQinBohuiLiuFnjingKongZhijunCheLiyuChen
Yuhng Zhng, Hiping Du, Tintin Zho, Chunmei Lio, Tu Feng, Jun Qin, Bohui Liu,Fnjing Kong, Zhijun Che*, Liyu Chen,*
a Guangdong Key Laboratory of Plant Adaptation and Molecular Design, Guangzhou Key Laboratory of Crop Gene Editing, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences, Guangzhou University, Guangzhou 510006, Guangdong, China
b Hebei Laboratory of Crop Genetics and Breeding, National Soybean Improvement Center Shijiazhuang Sub-Center, Huang-Huai-Hai Key Laboratory of Biology and Genetic Improvement of Soybean,Ministry of Agriculture and Rural Affairs,Institute of Cereal and Oil Crops,Hebei Academy of Agricultural and Forestry Sciences,Shijiazhuang 050035,Hebei,China
c School of Ecological Engineering, Guizhou University of Engineering Science, Bijie 551700, Guizhou, China
d School of Agriculture, Ningxia University, Yinchuan 750021, Ningxia, China
e College of Life Science, Hebei Normal University, Shijiazhuang 050024, Hebei, China
Keywords:GmTOC1b GmWRKY40 Soybean Soybean mosaic virus
ABSTRACT Soybean (Glycine max) is a major oil and feed crop worldwide.Soybean mosaic virus (SMV) is a globally occurring disease that severely reduces the yield and quality of soybean.Here,we characterized the role of the clock gene TIMING OF CAB EXPRESSION 1b (GmTOC1b) in the resistance of soybean to SMV.Homozygous Gmtoc1b mutants exhibited increased tolerance to SMV strain SC3 due to the activation of programmed cell death triggered by a hypersensitive response.Transcriptome deep sequencing and RT-qPCR analysis suggested that GmTOC1b likely regulates the expression of target genes involved in the salicylic acid(SA)signaling pathway.GmTOC1b binds to the promoter of GmWRKY40,which encodes a protein that activates the expression of SA-mediated defense-related genes.Moreover,we revealed that the GmTOC1bH1 haplotype, which confers increased tolerance to SMV, was artificially selected in improved cultivars from the Northern and Huang-Huai regions of China.Our results therefore identify a previously unknown SMV resistance component that could be deployed in the molecular breeding of soybean to enhance SMV resistance.
1.Introduction
Soybean mosaic virus (SMV) is a positive-sense single-stranded RNA virus belonging to the Potyvirus genus of the Potyviridae family that infects soybean plants worldwide [1].SMV causes symptoms such as dwarfism and leaf mosaicism, wrinkling, necrosis,and curling after infection [2,3].The total yield loss can reach 15%–50% under environmental conditions conducive to disease development [4].In severe cases, the yield loss can reach more than 70% or even 100%, thus restricting yields and quality in soybean [5].Twenty-two SMV strains (SC1–SC22) from China and 7 strains(G1–G7)from the United States have been identified based on differences in the pathogenicity and virulence of various SMV isolates [6,7].
Four independent loci, Rsv1, Rsv3, Rsv4, and Rsv5, confer resistance to diverse SMV strains in soybean.Rsv1 is composed of a cluster of multiple nucleotide-binding leucine-rich repeat (NB-LRR)genes,which encode typical resistance(R)proteins that are important for regulating disease resistance in plants [8].Rsv1 confers resistance to strains G1–G6 but does not protect again infection by G7 [9].The candidate gene for Rsv3 encodes a protein with a coiled-coil (CC) motif, a nucleotide-binding site (NBS), and an LRR domain;Rsv3 confers extreme resistance(ER) to specific strains of SMV, including G5, G6, G7, and G5H [10].Rsv4 encodes an RNase H family protein that degrades double-stranded RNA (dsRNA), a typical feature of plant virus genomes.Indeed, Rsv4 enters viral replication components to degrade the replication of viral dsRNA by interacting with the P3 protein of SMV [11].Rsc4-3 encodes a cell-wall localized NLR-type protein.Rsc4-3 directly interacts with the cylindrical inclusion (CI) protein in the apoplast to induce a hypersensitive response(HR),which confers tolerance to SMV[5].
Many WRKY transcription factors participate in plant defense responses [12–16].In oilseed rape (Brassica napus), BnWRKY33 positively regulates resistance to the pathogenic fungus Sclerotinia sclerotiorum by enhancing the expression of genes involved in camalexin biosynthesis and genes regulated by salicylic acid (SA)and jasmonic acid (JA).By contrast, overexpression of BnWRKY15 increased susceptibility to S.sclerotiorum by inhibiting the transcriptional activation of BnWRKY33 [17].Similarly, GhWRKY41 homodimerizes and activates the expression of CINNAMATE 4-HYDROXYLASE (GhC4H) and 4-COUMARATE-CoA LIGASE (Gh4CL)to increase the accumulation of lignin and flavonoids in upland cotton (Gossypium hirsutum), which enhances resistance against the fungus Verticillium dahliae[18].CaWRKY27 positively regulates the resistance of pepper (Capsicum annuum) to the bacterium Ralstonia solanacearum by upregulating the expression of genes associated with HR, SA, JA, and ethylene signaling [19].In soybean,GmWRKY12 confers tolerance to drought and salt stress, likely by increasing the contents of proline (Pro) and malondialdehyde(MDA)[20].GmWRKY27 also improves salt and drought tolerance and interacts with GmMYB174.The resulting heterodimer binds to the promoter region of the NAC family member GmNAC29 to reduce its expression[21].Furthermore,GmWRKY13,GmWRKY21,and GmWRKY54 play important roles in plant responses to abiotic stress [22].
Light plays a crucial role in regulating the balance between soybean growth and defense.Soybean plants infected with SMV exhibited varying degrees of disease phenotypes under different light intensities.The expression levels of genes involved in plantpathogen interactions, mitogen-activated protein kinase (MAPK)signaling pathways,and SA-mediated defense pathways were also affected by light intensity [23].Temperature also has important effects on the phenotypes induced by SMV:20–25°C is the optimal temperature for viral replication and spreading, whereas disease symptoms are suppressed between 26 and 28 °C.However, at 29.5 °C, the virus is still present in the plant but the symptoms are very mild[24].Upon sensing changes in external light and temperature conditions, signals are transmitted to the plant circadian clock, which responds by regulating the transcription of downstream genes, protein activity, or metabolism to adjust the adaptability of plants to external stimuli [25].Therefore, the circadian clock might be involved in regulating the resistance of soybean to SMV.Indeed, a close relationship exists between the circadian clock and plant immunity[57,58].TOC1 serves as the core component of the circadian clock.However,whether TOC1 participates in the plant immune response in soybean is currently unknown.
In this study,we analyzed the role of GmTOC1b in the resistance of soybean to SMV and the potential underlying regulatory mechanisms.We established that GmTOC1b indirectly regulates the expression of defense-related genes mediated by SA by directly inhibiting GmWRKY40 expression, which ultimately results in reduced SMV resistance of the host plant.Additionally,we investigated the natural variation at GmTOC1b from a diversity panel of 2479 soybean accessions.By integrating functional validation of haplotypes with phenotypic data obtained from soybean accessions following inoculation with SMV, we revealed the selection pattern of GmTOC1b haplotypes in improved soybean cultivars.Our findings provide insight into the regulatory network affected by SMV and lay the foundation for breeding SMV-resistant soybean lines.
2.Materials and methods
2.1.Plant materials and phenotyping assays
The soybean cultivar Williams 82 (W82) and the homozygous Gmtoc1b mutants Gmtoc1b-1 (with a 7-bp deletion in GmTOC1b)and Gmtoc1b-2(with a 4-bp deletion in GmTOC1b)obtained previously[35]were used for phenotyping assays and further functional analysis.Leaves collected from soybean cultivar Nannong 1138–2 infected with SMV strain SC3 were used in this study.
The inoculation of soybean with SMV was performed as previously described with minor modifications [26].Typical fresh diseased leaves infected with SC3 from highly susceptible variety Nannong 1138–2 were harvested and cut into pieces.The leaves were homogenized at a ratio of 1 g leaf tissue to 20 mL 0.1 mol L-1phosphate buffer (pH 7.0) under ice-cold conditions.The virus inoculum was generated by filtering the homogenate through gauze.Plant materials were cultivated in an illuminating incubator at 25 °C under a 16 h light/8 h dark photoperiod.When unifoliate leaves were fully expanded, the virus inoculum mixed with carborundum powder was gently rubbed onto the leaves with a brush.Leaves rubbed with 0.1 mol L-1phosphate buffer (pH 7.0) mixed with carborundum powder were used as the un-inoculated control.All rubbed leaves were briefly rinsed with sterile water after 5 min.Disease symptoms were evaluated at 21 days post inoculation(dpi).
2.2.3,3′-Diaminobenzidine (DAB) and trypan blue staining
DAB staining was performed as previously described [27].Briefly, 50 mg DAB tetrahydrochloride hydrate was dissolved in 50 mL sterilized H2O and mixed with 25 μL Tween 20 to generate 1 mg mL-1DAB solution.Leaves were immersed into the DAB solution and covered with aluminum foil to avoid light exposure.The leaves were then gently vacuum-infiltrated for 10 min to ensure even penetration of the DAB solution into the leaves.After overnight incubation, the DAB staining solution was removed and replaced with bleaching solution (ethanol: acetic acid: glycerol = 3:1:1, v/v/v); the samples were placed in a boiling water bath for approximately 15 min to remove chlorophyll.The bleaching solution was replaced with fresh bleaching solution and left to stand for 30 min.Finally, photographs were taken on a black background.
Trypan blue staining was conducted as previously described[5].Briefly, leaves were immersed in trypan blue staining solution(20 mL absolute ethanol,5 mL lactic acid,5 mL phenol,5 mL glycerol, 5 mL sterile water, and 10 mg trypan blue) and vacuuminfiltrated for 10 min.The samples were heated in a boiling water bath for 10 min and incubated overnight at room temperature.The leaves were destained three times in chloral hydrate (2.5 g mL-1).Finally, photographs were captured under a Zeiss Axio Imager A2 microscope (Zeiss, Oberkochen, Germany).
2.3.RNA-seq analysis
Leaves from W82 and Gmtoc1b-2 mutants inoculated with SMV or mock-inoculated with 0.1 mol L-1phosphate buffer (pH 7.0)were collected at 14 dpi.Total RNA was extracted from the samples and enriched for mRNA with Oligo(dT)beads.The enriched mRNA was fragmented into short fragments for first-strand cDNA synthesis.The purified cDNA fragments were ligated with sequencing adapters and the cDNA library was sequenced using an Illumina Novaseq6000 instrument by Gene Denovo Biotechnology Co.(Guangzhou, Guangdong, China).The clean reads were mapped to the soybean reference genome(Wm82.a4.v1)using HISAT2.2.4[28].Differential expression analysis was performed using DESeq2 and edgeR software [29,30].The differentially expressed genes(DEGs)were chosen based on the threshold value of a false discovery rate(FDR)<0.05 and absolute fold-change ≥2.The DEGs were submitted to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for GO term and pathway enrichment analyses [31,32].The RNA-seq data were deposited at the NCBI Sequence Red Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra; accessed on May 10, 2023) under accession number PRJNA957910.
2.4.RNA extraction and RT-qPCR analysis
Total RNA was extracted from the samples using a FastPure Plant Total RNA Isolation Kit according to the manufacturer’s instructions (Vazyme, Nanjing, Jiangsu, China).First-strand cDNA synthesis was performed with a HiScript III 1st Strand cDNA Synthesis Kit(Vazyme).qPCR was conducted on a Roche LightCycle480 system with AceQ Universal SYBR qPCR Master Mix (Vazyme).All experiments were conducted with three biological replicates.GmEF1b (ELONGATION FACTOR 1b, Glyma.02G276600) and NbActin were used as reference transcripts.
2.5.Immunoblot and chromatin immunoprecipitation-qPCR analyses
Total proteins were extracted from the leaves of transgenic plants overexpressing GmTOC1b or GmWRKY40.The samples were ground into a powder and mixed thoroughly with protein extraction buffer.Following centrifugation at 4 °C for 10 min at 12,000 r min-1,the supernatant was transferred to a fresh tube and mixed with 5× SDS loading buffer before being boiled in a boiling water bath for 10 min.Total soluble proteins were separated by SDSPAGE and then transferred to a PVDF membrane for immunoblot analysis, which was detected using anti-FLAG or anti-Actin antibodies as described previously [33].
For the ChIP-qPCR assay,leaf samples were collected from W82 and GmTOC1b-OE materials at 14 dpi with SMV inoculum; the experiment was essentially performed as previously described[34].The ground samples were mixed with ChIP extraction buffer and fixed with formaldehyde.Glycine was then added to terminate the reaction.Lysis buffer was added to release the protein-DNA complex from cells.Cell and solubilized protein debris were removed by centrifugation, and the protein-DNA complexes were sonicated to break into fragments of approximately 500 bp.The solubilized chromatin complexes were immunoprecipitated with affinity anti-Flag antibody coupled with magnetic beads (SIGMA,Darmstadt, Germany).The co-immunoprecipitated DNA was eluted and purified for qPCR analysis.Relative fold enrichment was evaluated by normalizing the amount of target DNA fragment relative to the genomic fragment of the reference gene GmEF1b and then by normalizing the value of the input DNA.
2.6.Subcellular localization of GmWRKY40
The full-length GmWRKY40 (Glyma.13G370100) coding sequence without the stop codon was cloned in-frame and upstream of the green fluorescent protein (GFP) sequence driven by the cauliflower mosaic virus (CaMV) 35S promoter.The resulting plasmid was transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101, which was then infiltrated into Nicotiana benthamiana leaves.The plants were grown at 26 °C under a 16 h light/8 h dark photoperiod for 48 h.GFP fluorescence was observed under a Zeiss LSM 800 confocal laser scanning microscope (Zeiss, Oberkochen, Germany).N.benthamiana leaves were infiltrated with an Agrobacterium culture carrying the p35S:GFP plasmid as an expression control.
2.7.Transient expression assay
To generate promoter-driven firefly luciferase (LUC) reporter constructs,~2-kb GmWRKY40 promoter fragments were amplified from W82 genomic DNA;the PCR products were purified and individually cloned into the pGreenII 0800-LUC vector.The resulting pGmWRKY40:LUC constructs were used as reporters, with pGmUBI:3 × FLAG and pGmUBI:GmTOC1bH1/H2/H3-3 × FLAG used as effectors.The constructs pGmUBI:GmTOC1bH1/H2/H3-3 × FLAG and pGmWRKY40:LUC were co-infiltrated into N.benthamiana leaves.The constructs pGmUBI:3 × FLAG and pGmWRKY40-LUC were coinfiltrated into N.benthamiana leaves as the negative control.Relative LUC activity was determined by measuring LUC and Renilla luciferase (REN) activity using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) on a Biotek Synergy H1 Microplate Reader (Agilent, Santa Clara, CA, USA).Three independent biological replicates were performed in this assay.Relative LUC activity was calculated as the ratio between LUC and REN activities.Digital images were taken after spraying 1 mmol L-1of D-Luciferin sodium salt onto infiltrated N.benthamiana leaves(Sangon Biotech, Shanghai, China).
2.8.Statistical analyses
For RT-qPCR analysis, at least three individual plants were pooled in each sample and three independent technical replicates were performed.All data were presented as mean values ± standard error (SE).Data were analyzed with GraphPad Prism 9 software.Significance levels of differences were calculated by one-tailed,two sample Student’s t-tests and indicated by different lowercase letters in the graphs.
3.Results
3.1.Knockout of GmTOC1b enhances SMV resistance
We previously generated two homozygous Gmtoc1b mutants by genome editing to knock out GmTOC1b in the W82 background[35].To investigate a possible role for GmTOC1b in soybean host resistance to SMV, we inoculated fully expanded unifoliate leaves with SMV and evaluated the phenotypes of Gmtoc1b mutants and wild-type W82 at 21 dpi.We observed more pronounced necrosis symptoms on the upper un-inoculated leaves of the Gmtoc1b mutants than in W82 after inoculation with SMV, which is indicative of greater resistance mediated by HR.Moreover,the top leaves of the Gmtoc1b mutants exhibited milder deformity and shrinkage symptoms compared to W82 (Figs.1A, S1).We assessed the expression of the CP (encoding the SMV coat protein) gene in the Gmtoc1b mutants and W82, which indicated that CP is expressed at much lower levels in the Gmtoc1b mutants relative to W82(Fig.1B).Together, these data reveal that knocking out GmTOC1b enhances the resistance of soybean to SMV.
3.2.Knocking out GmTOC1b promotes programmed cell death
During the plant immune response,H2O2is produced at the site of infection, which acts as a key signaling molecule to inhibit pathogen growth and limits the spread of pathogens to nearby cells by inducing HR and causing the localized programmed cell death of infected cells.To investigate whether GmTOC1b affects H2O2levels, we assessed the accumulation of H2O2by 3,3′-diaminobenzidine(DAB)staining.We observed no clear difference between upper leaves of the mock-inoculated Gmtoc1b mutants and W82.However, we detected darker brown staining in the leaves of Gmtoc1b compared to W82 plants following inoculation with SMV (Fig.2A).Because stronger necrosis symptoms developed on the leaves of the Gmtoc1b mutants after inoculation, we reasoned that GmTOC1b might also play a role in the HR.Consistent with this hypothesis, Gmtoc1b leaves showed more extensive staining with trypan blue compared to W82 leaves following inoculation (Fig.2B).These results suggest that the resistance to SMV observed in the Gmtoc1b mutants might be due to programmed cell death triggered by the HR.
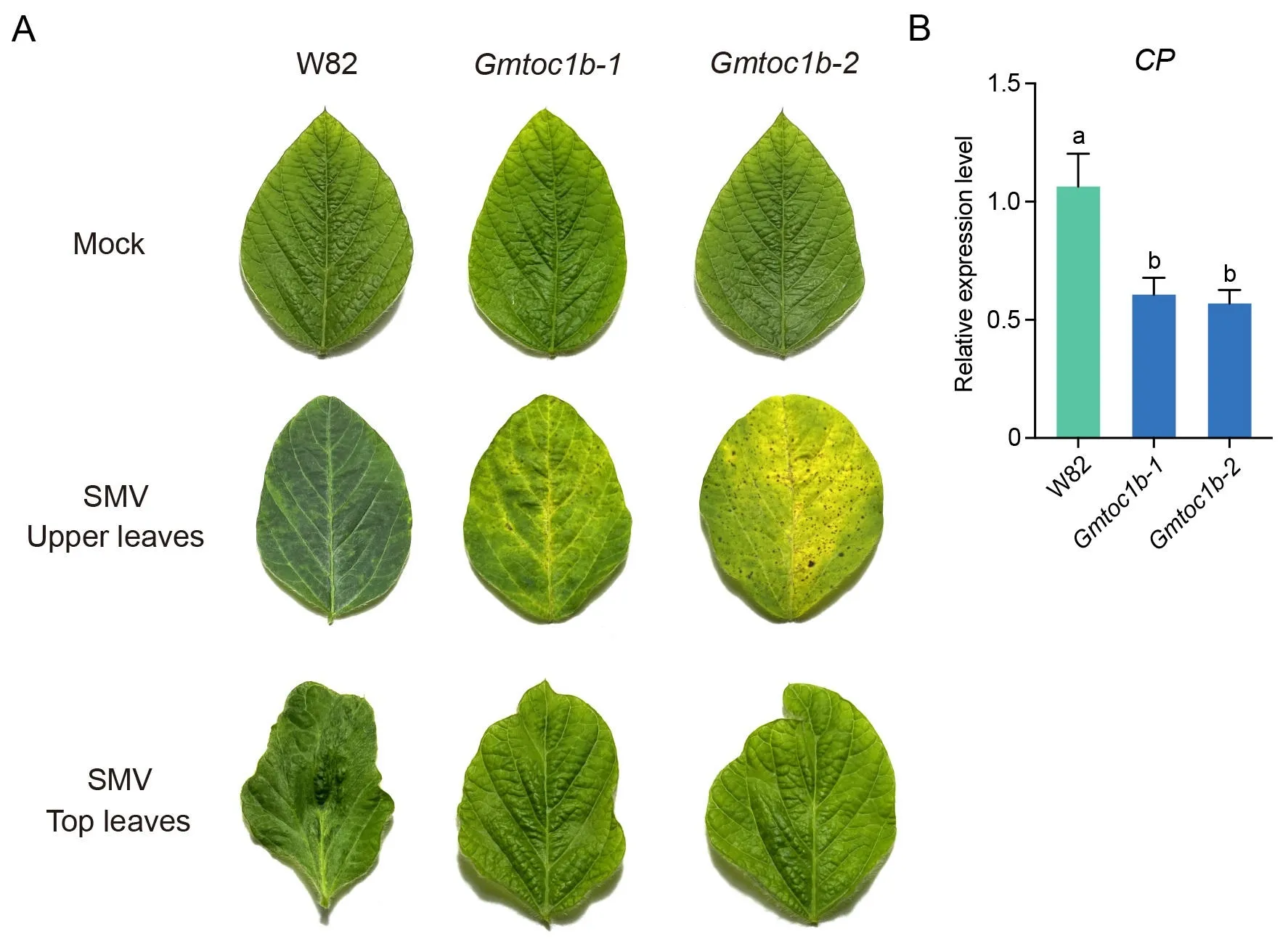
Fig.1.Knockout of GmTOC1b enhances SMV tolerance in soybean.(A)Phenotypes of the upper leaves and top leaves from the Gmtoc1b mutants and wild-type W82 at 21 dpi with SMV or 0.1 mol L-1 phosphate buffer(Mock control).(B)RT-qPCR analysis of the accumulation of SMV CP transcripts after inoculation with SMV.CP gene expression was normalized to that of the reference gene GmEF1b.Each experiment was performed with three biological replicates.Significant differences were indicated by different lowercase letters, as determined by Student’s t-test.dpi, day post inoculation.
3.3.Differentially expressed genes define signaling pathways regulated by GmTOC1b under SMV inoculation
To identify the downstream target genes regulated by GmTOC1b during SMV inoculation, we conducted RNA-seq analysis using leaves from Gmtoc1b-2 mutant and W82 plants that were mock-inoculated or inoculated with SMV.Over 92% of the clean reads uniquely aligned to the W82 reference genome.Pearson’s correlation analysis revealed a high degree of correlation among the three biological replicates for each sample,supporting the reliability of the RNA-seq datasets.We identified differentially expressed genes (DEGs) using an absolute fold-change ≥2 and a false discovery rate (FDR) < 0.05.We obtained 6989 DEGs (4824 upregulated and 2165 downregulated) from the W82-SMV vs.W82-mock comparison; 10,174 DEGs (6792 upregulated and 3382 downregulated) from the Gmtoc1b-SMV vs.Gmtoc1b-mock comparison; 4339 DEGs (2112 upregulated and 2227 downregulated) from the Gmtoc1b-mock vs.W82-mock comparison; and 8446 DEGs (4425 upregulated and 4021 downregulated) from the Gmtoc1b-SMV vs.W82-SMV comparison (Fig.3A).Furthermore,714 DEGs from Gmtoc1b-mock vs.W82-mock and Gmtoc1b-SMV vs.W82-SMV were shared with the W82-SMV vs.W82-mock comparison (which are considered to be SMV-responsive genes)(Fig.3B).These results indicate that the loss of GmTOC1b function has a profound influence on the soybean SMV-response network.
We looked at the DEGs from the Gmtoc1b-mock vs.W82-mock and Gmtoc1b-SMV vs.W82-SMV comparisons in more detail.We observed similar gene expression patterns among the three biological replicates for each genotype, but W82 and Gmtoc1b showed clear differences (Fig.3C, D).To better understand the biological functions of GmTOC1b,we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the DEGs derived from the W82 and Gmtoc1b comparisons under mock treatment or inoculation with SMV.We detected a significant enrichment for DEGs involved in plant hormone signal transduction in the two comparisons: Gmtoc1b-mock vs.W82-mock and Gmtoc1b-SMV vs.W82-SMV.Moreover, DEGs involved in plant–pathogen interactions and the MAPK signaling pathway were significantly enriched in the Gmtoc1b-SMV vs.W82-SMV comparison.We also noticed the enrichment of DEGs among metabolic pathways related to plant growth and development, including starch and sucrose metabolism, circadian rhythms, flavonoid biosynthesis, ABC transporters, and nitrogen metabolism (Fig.3E, F).These results suggest that GmTOC1b regulates the expression of genes that participate in these pathways to influence the SMV response of soybean.
3.4.GmTOC1b regulates the expression of salicylic acid signaling and defense-related genes
SA is an important plant hormone in plant defense responses.SA accumulation triggers a series of immune responses, such as the expression of disease resistance genes,cell wall strengthening,and the production of secondary metabolites and antimicrobial proteins.During infection by SMV, SA strongly accumulates in leaves,thereby activating the plant immune response and enhancing plant resistance against the invading pathogen.Through gene functional annotation,we noticed the presence of at least 17 DEGs from the Gmtoc1b-SMV vs.W82-SMV comparison associated with plant–pathogen interactions and plant hormone signal transduction related to SA signaling.These genes included NONEXPRESSER OF PATHOGENESIS RELATED GENES 1 (NPR1), ENHANCED DISEASE SUSCEPTIBILITY 1(EDS1),PHYTOALEXIN DEFICIENT 4(PAD4),PATHOGENESIS RELATED 1(PR1),PR2,and PR10.Additionally,we detected the upregulation of three RESPIRATORY BURST OXIDASE HOMOLOGS(RBOH) genes in SMV-inoculated Gmtoc1b compared to inoculated W82; these genes encode NADPH-oxidases that produce reactive oxygen species.WRKY40 (Glyma.13G370100) was also significantly upregulated specifically in Gmtoc1b following inoculation with SMV(Fig.4A).We validated the expression of several genes by RTqPCR(Fig.4B–I).We also quantified SA and SAG(Salicylic acid 2-Oβ-glucoside) levels in the upper un-inoculated leaves of the Gmtoc1b mutants and W82 following inoculation with SMV,revealing that Gmtoc1b mutants accumulated significantly higher level of SA and SAG following inoculation with SMV (Fig.4J, K).These results support the notion that GmTOC1b likely regulates SA signaling to modulate the plant response to SMV.
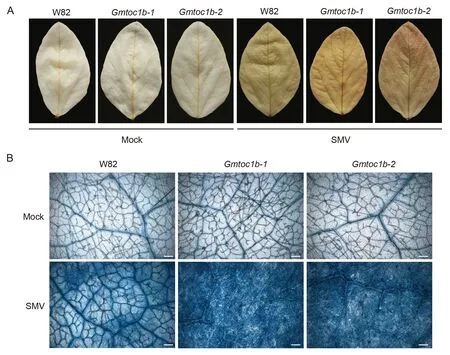
Fig.2.GmTOC1b modulates programmed cell death in response to SMV.(A)H2O2 levels in the upper leaves of the Gmtoc1b mutants and W82 at 14 dpi with SMV or 0.1 mol L-1 phosphate buffer (Mock control), as detected by DAB staining.(B) Cell death in the upper leaves of the Gmtoc1b mutants and W82 at 14 dpi with SMV or 0.1 mol L-1 phosphate buffer (Mock control), as observed by trypan blue staining and imaging under a Zeiss Axio Imager A2 microscope.Scale bars, 100 μm.dpi, day post inoculation.
3.5.GmTOC1b binds to the promoter of GmWRKY40
Previous chromatin immunoprecipitation followed by deep sequencing(ChIP-seq)analysis showed that Arabidopsis(Arabidopsis thaliana) TOC1 directly binds to the region of WRKY40(At1g80840) gene [36].We speculated that GmTOC1b might also bind to the GmWRKY40 in soybean.We analyzed the 2-kb promoter fragment of the GmWRKY40 genes, which is upregulated in Gmtoc1b, to identify the cognate cis-elements bound by TOC1.Indeed, we detected one typical morning element (ME) and three typical hormone up-regulated at dawn motifs (HUD) in the GmWRKY40 promoter.We collected leaf samples from transgenic soybean plants overexpressing GmTOC1b-3 × FLAG (GmTOC1b-OE)and measured GmTOC1b transcript levels by RT-qPCR and GmTOC1b-3 × FLAG abundance by immunoblotting with anti-FLAG antibody.GmTOC1b expression level were 4- to 28-fold higher in the GmTOC1b-OE lines compared to W82; we detected FLAG-tagged GmTOC1b in these OE lines, but not in W82(Fig.5A, B).Having confirmed the accumulation of GmTOC1b-3 × FLAG in the GmTOC1b-OE lines, we collected the upper leaves from GmTOC1b-OE#3 following inoculation with SMV for a ChIPqPCR assay.We determined that GmTOC1b binds to two regions(P2 and P4)in the GmWRKY40 promoter(Fig.5C).We conclude that GmTOC1b binds to the GmWRKY40 promoter.
3.6.GmWRKY40 represses the expression of PR genes
Identifying the subcellular distribution of GmWRKY40 is essential for predicting its function.Accordingly, we infiltrated a construct harboring GmWRKY40-GFP (encoding a fusion between GmWRKY40 and green fluorescent protein) under the control of the 35S promoter into N.benthamiana leaves;we detected GFP fluorescence specifically in the nucleus, whereas the signal observed in leaves infiltrated with p35S:GFP was distributed evenly throughout the cells.These results indicate that GmWRKY40 is a nucleuslocalized protein (Fig.5D).
We also constructed the pGmUBI:GmWRKY40-3 ×FLAG overexpression vector,which we infiltrated into N.benthamiana leaves via Agrobacterium-mediated infiltration, using the empty vector as a negative control.Following infiltration, we grew the plants at 26 °C under a 16 h light/8 h dark photoperiod for 48 h before collecting the infiltrated leaves for RT-qPCR and immunoblotting analysis.GmWRKY40 expression were significantly higher in leaves transiently expressing GmWRKY40 relative to the empty vector control;we also detected FLAG-tagged GmWRKY40 in the overexpression lines, but not in the empty vector control (Fig.5E, F).Using the same RNA samples, we measured the transcript abundance of NbPR1a,NbPR2,and NbPR4.These genes were significantly upregulated in leaves transiently expressing GmWRKY40, indicating that GmWRKY40 activates the expression of defense-related genes involved in the SA pathway (Fig.5G–I).
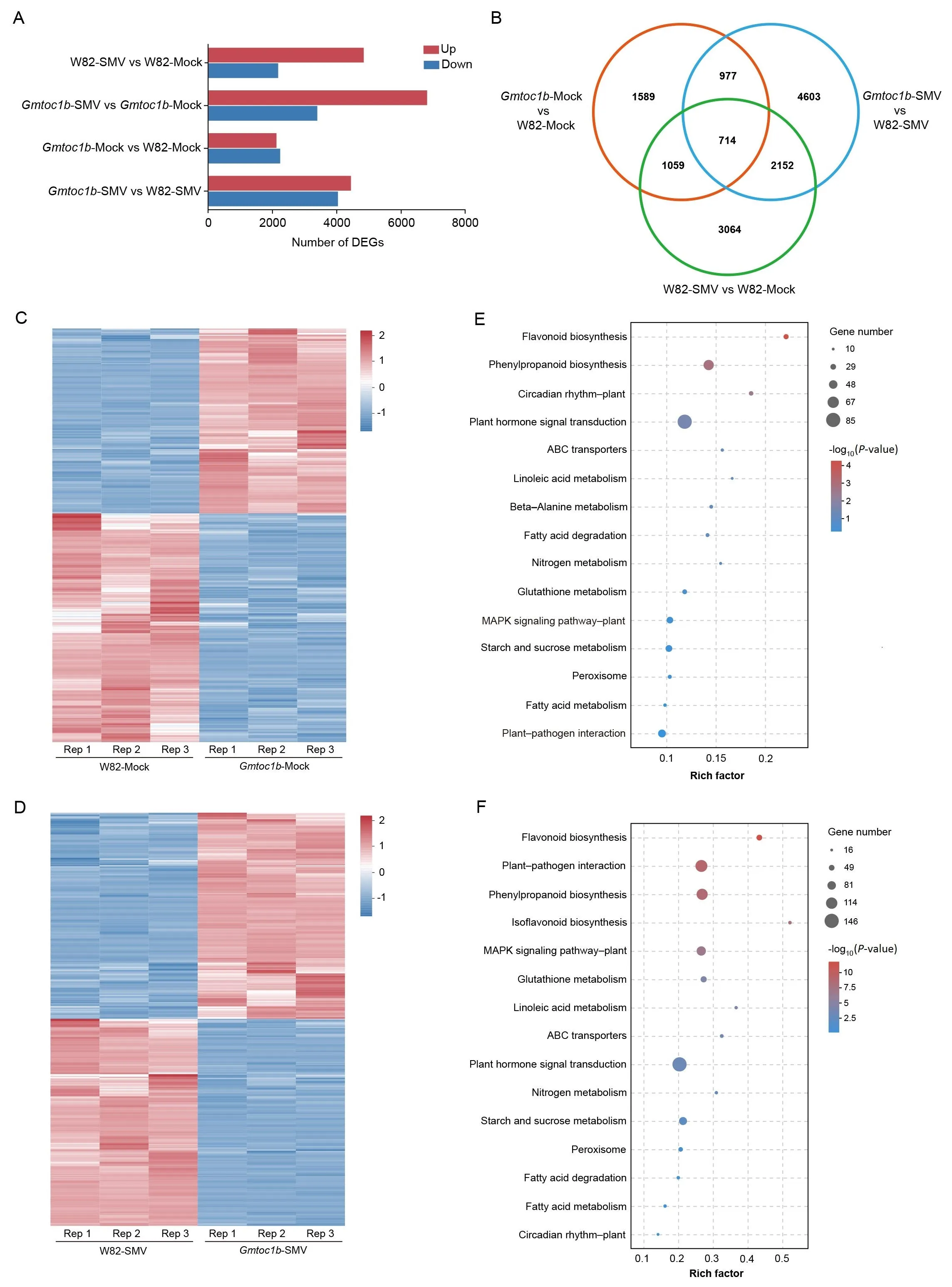
Fig.3.Identification of signaling pathways enriched in the DEGs based on RNA-seq analysis.(A) Number of upregulated (Up) and downregulated (Down) differentially expressed genes(DEGs)from the four comparisons.(B)Venn diagram showing the extent of overlap between DEGs from the Gmtoc1b-Mock vs.W82-Mock,Gmtoc1b-SMV vs.W82-SMV and W82-SMV vs.W82-Mock comparisons.(C)Heatmap representation of the normalized expression levels of the DEGs in W82 and Gmtoc1b mutants following inoculation with 0.1 mol L-1 phosphate buffer(Mock control).Three biological replicates were analyzed per group.The blue to red scale bar indicates FPKM values from low to high.(D) Heatmap representation of normalized expression levels of the DEGs in W82 and the Gmtoc1b mutant inoculated with SMV.Three biological replicates were analyzed per group.(E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the DEGs identified between W82 and the Gmtoc1b mutant following inoculation with 0.1 mol L-1 phosphate buffer (Mock control).Rich factor represents the ratio of the number of DEGs enriched in the pathway relative to the number of all genes involved in the pathway.Circle size represents the number of enriched DEGs,and the color indicates the–log10(P-value).(F)KEGG pathway enrichment analysis of the DEGs identified between in W82 and the Gmtoc1b mutant following inoculation with SMV.

Fig.4.RT-qPCR validation of the DEGs in W82 and the Gmtoc1b mutants.(A)Heatmap representation of the 21 DEGs specifically induced in the Gmtoc1b mutant following inoculation with SMV.The blue to red scale bar indicates normalized FPKM values from low to high.(B–I) Validation of the transcript levels of selected DEGs in W82 and Gmtoc1b mutant following inoculation with SMV or 0.1 mol L-1 phosphate buffer(Mock control).The relative expression level was adjusted to the reference gene GmEF1b for normalization.(J–K)SA and SAG(Salicylic acid 2-O-β-glucoside)levels in the upper un-inoculated leaves of Gmtoc1b mutant and W82 at 21 dpi with SMV.Each experiment was performed with three biological replicates.Significant differences were indicated by different lowercase letters, as determined by Student’s t-test.dpi, day post inoculation.
3.7.Artificial selection of the GmTOC1bH1 haplotype in improved soybean cultivars from Northern region and the Huang-Huai region in China
We explored the natural variation of GmTOC1b using genome resequencing data from a diversity panel of 2479 soybean accessions, including 1280 improved cultivars, 671 landraces and 528 wild accessions collected from China [37–43], leading to the discovery of three major haplotypes: GmTOC1bH1, GmTOC1bH2, and GmTOC1bH3(Fig.6A).To test for functional differences among these three haplotypes,we performed a transient dual-luciferase expression assay using the GmWRKY40 promoter cloned upstream of the firefly luciferase (LUC) reporter gene together with three effector constructs, each harboring a different GmTOC1b haplotype driven by the soybean Ubiquitin (GmUBI) promoter (Fig.6B).Coinfiltration of N.benthamiana leaves with the LUC reporter construct and each effector construct revealed that the GmTOC1bH3haplotype most strongly repressed relative LUC activity driven by the GmWRKY40 promoter, followed by GmTOC1bH2, with GmTOC1bH1showing the weakest inhibitory effect (Fig.6C, D).

Fig.5.GmTOC1b binds to the promoter of GmWRKY40 and indirectly repressing the expression of PR genes.(A)Relative GmTOC1b transcript levels in GmTOC1b-OE and W82.The relative expression level was adjusted to the reference gene GmEF1b for normalization.(B)Immunoblot analysis of GmTOC1b-3FLAG abundance in W82 and GmTOC1b-OE.An anti-FLAG antibody was used to recognize GmTOC1b-3FLAG; GmActin was used as the loading control.(C) Distribution of TOC1 binding motifs in the GmWRKY40 promoter(top)and enrichment analysis via ChIP-qPCR(bottom).(D)Subcellular localization of GmWRKY40.GFP,green fluorescent protein;BF,brightfield;Merge,GFP,DAPI and BF images.Scale bars, 15 μm.(E) Immunoblot analysis of GmWRKY40 abundance in N.benthamiana leaves transiently expressing the empty vector control and GmWRKY40-OE.An anti-FLAG antibody was used to recognize GmWRKY40-3FLAG; NbActin was used as the loading control.(F) Relative GmWRKY40 transcript levels in N.benthamiana leaves transiently expressing GmWRKY40 or the empty vector control.Relative expression level was normalized to that of the reference gene NbActin.(G–I)Relative transcript levels of NbPR1a,NbPR2,and NbPR4 in N.benthamiana leaves collected from plants 48 h after transient infiltration with the indicated constructs.Relative expression level was normalized to that of the reference gene NbActin.dpi, day post inoculation.
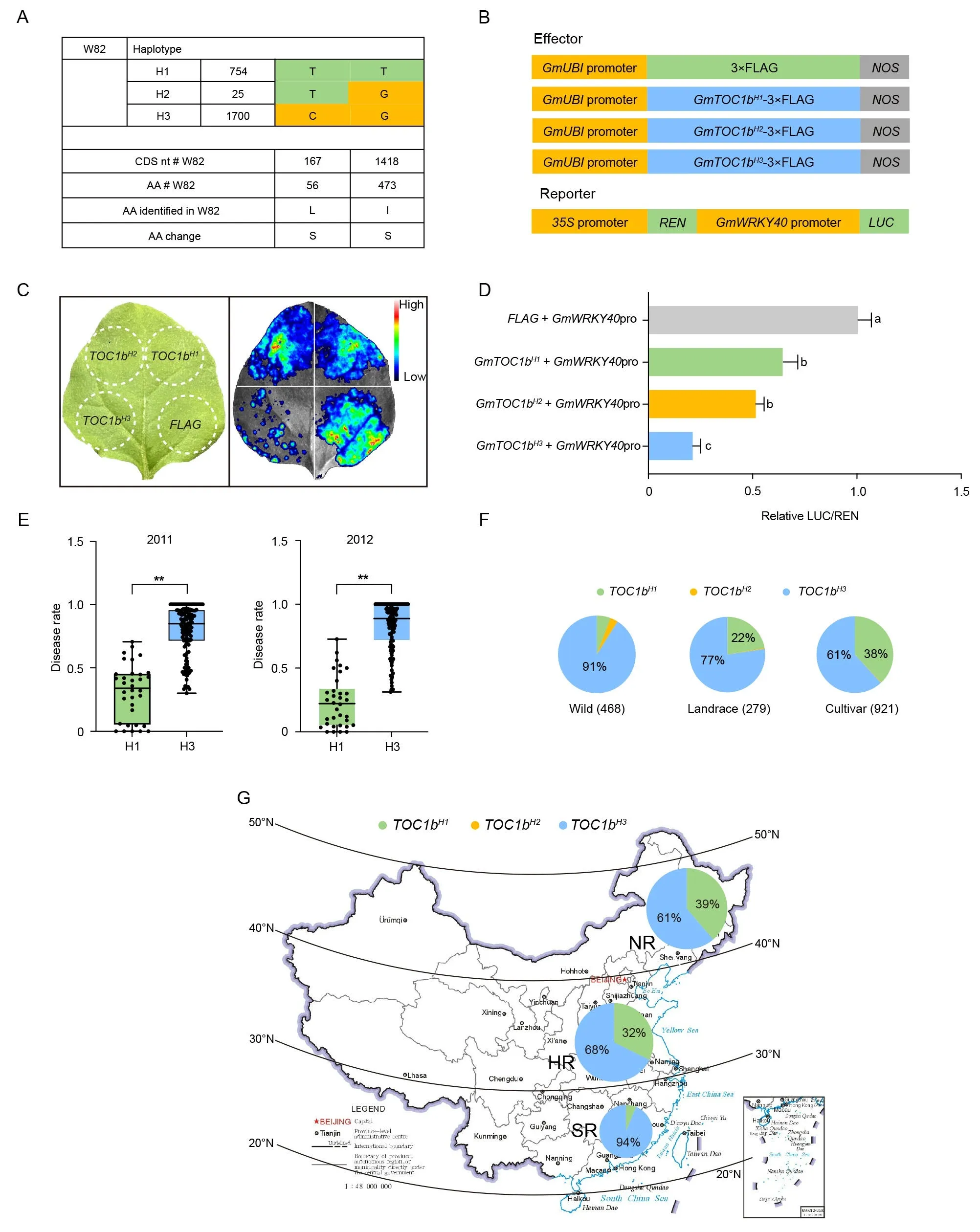
Fig.6.Functional analysis and distribution of GmTOC1b haplotypes.(A)Haplotypes of the GmTOC1b gene from a 2479-variety panel.The variants were annotated based on the Williams 82 a4.v1 reference genome.(B)Diagram of the constructs used in the transient co-infiltration assay.(C)Image illustrating firefly luciferase(LUC)activity driven by the GmWRKY40 promoter as a function of the GmTOC1b haplotype.(D) Relative luciferase activity derived from the pGmWRKY40:LUC reporter construct upon coinfiltration with various effector constructs expressing different GmTOC1b haplotypes.Relative LUC activity was normalized to 35S promoter-driven Renilla luciferase (REN)activity.(E) Disease rates of a 219-accession diversity panel with the GmTOC1bH1 or GmTOC1bH3 haplotypes grown in Nanjing over two years.(F) Distribution of GmTOC1b haplotypes in wild soybeans, landraces, and cultivars.(G) Geographical distribution of GmTOC1b haplotypes among improved cultivars in China.NR, northern region; HR,Huang-Huai region; SR, southern region.The map was downloaded at the website https://bzdt.ch.mnr.gov.cn, and the map content approval number is GS (2019) 1673.
To examine the phenotypic consequences of the GmTOC1b haplotypes,we analyzed the differences in disease rate following inoculation of 219 soybean accessions with SMV strain SC3 reported previously[43].The GmTOC1bH1haplotype conferred increased tolerance of soybean to SMV over the two years examined (Fig.6E).We also detected the distribution of each of the three haplotypes in the wild soybeans,landraces,and improved cultivars,suggesting that GmTOC1bH3might be the ancestral haplotype and accounted for 91% of wild soybeans.Furthermore, the proportion of GmTOC1bH1in landraces and improved cultivars gradually increased compared to wild soybeans and accounted for 22% and 38%, respectively.Finally, we investigated the distribution of the haplotypes among improved cultivars from the Northern region,the Huang-huai region, and the Southern region of China.Except for the GmTOC1bH1haplotype, which accounted for 6% of all cultivars from the Southern region; the proportion of the GmTOC1bH1haplotype reached 39% and 32% in the improved cultivars from the Northern region and the Huang-Huai region of China, respectively.These results support the conclusion that the GmTOC1bH1haplotype, which enhances the tolerance of soybean to SMV, was artificially selected during the improvement of soybean cultivars from the Northern region and the Huang-Huai region in China.
4.Discussion
The circadian clock is involved in almost all aspects of plant growth, development, and responses to external and internal changes [44–48].In Arabidopsis, TOC1 directly binds to the promoter of the tricarboxylic acid cycle-related gene FUMARASE 2 to repress its expression at night,contributing to the balance of plant metabolism [49].TOC1 also binds to the promoter of the abscisic acid (ABA)-related gene ABA-BINDING PROTEIN (ABAR) to mediate drought tolerance[50].In the crop species rice(Oryza sativa),OsTOC1 regulates tillering and panicle development by repressing CIRCADIAN CLOCK ASSOCIATED 1 (OsCCA1), thus affecting plant architecture and yield[51].However, little is known about the function of TOC1 in soybean.We previously demonstrated that GmTOC1b inhibits nodulation by suppressing the expression of the nodulationrelated genes NODULE INCEPTION 2a(GmNIN2a)and EARLY NODULIN 40–1(GmENOD40-1)[35].In this study,we characterized the function of GmTOC1b in the resistance of the soybean to SMV.Gmtoc1b mutants showed increased tolerance to SMV strain SC3.The programmed cell death triggered by HR was also more pronounced in the Gmtoc1b mutants.RNA-seq and RT-qPCR analyses suggested that target genes involved in the SA signaling pathway are significantly upregulated post-inoculation.SA plays an important role in the resistance of soybean to SMV [52–56].Three defense-related PR1 genes in soybean were reported to be specifically expressed in necrotic leaves after inoculation with SMV.The exogenous application of SA, but not methyl jasmonate (MeJA), on healthy leaves induced the expression of these three genes,indicating that GmPR1 genes are activated by SA in soybean [26].Moreover, knockout of
MALECTIN-LIKE RECEPTOR KINASE 1 (GmMLRK1) significantly reduced the levels of free SA and resistance to SMV strains SC7 and SC3, along with lower H2O2levels and programmed cell death in the mutants [43].Collectively, these results suggest that the GmTOC1b-regulated SA signaling pathway might be a primary factor affecting the resistance of soybean to SMV.
Previous studies revealed a key functional link between the circadian clock and plant immunity.Mutations in the core clock component LUX ARRHYTHMO(LUX)disrupt the circadian regulation of stomatal opening and closure.LUX also binds to the promoter of
EDS1 and JASMONATE-ZIM-DOMAIN PROTEIN 5 (JAZ5), likely acting through these genes to affect SA and JA signaling,which ultimately leads to broad-spectrum disease resistance to pathogens [57].Moreover,many R genes that mediate resistance against Hyaloperonospora Arabidopsis(Hpa)are regulated by the circadian regulator CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) [58].Due to the complex regulation relationship of transcription-translation feedback loops(TTFLs)involving TOC1 and other circadian clock components such as LUX and CCA1,whether GmTOC1b also affects the resistance of soybean to SMV by regulates circadian clock components or other defense-related pathways is currently unknown.
TOC1 directly binds to DNA, functioning as a transcriptional repressor to inhibit the expression of its target genes[59].Following inoculation with SMV, GmWRKY40 was expressed at significantly higher levels in the Gmtoc1b mutant compared to W82.We discovered two peaks in the Arabidopsis WRKY40 region as being significantly enriched for TOC1 binding in previously published ChIP-seq datasets [36].Therefore, we reasoned that GmWRKY40 is a likely target gene regulated by GmTOC1b.We confirmed this hypothesis by ChIP-qPCR and transient expression assays, establishing that GmTOC1b can binds to the GmWRKY40 promoter to repress its transcription.
GmWRKY40 expression is strongly induced following infection with Phytophthora sojae or the exogenous application of phytohormones such as SA, ethylene, MeJA, or ABA.GmWRKY40 facilitates ROS accumulation and stimulates the expression of marker genes such as GmPR1 in the SA signaling pathway, thereby increasing the resistance of soybean to Phytophthora sojae [60,61].Here we explored the function of GmWRKY40 and confirmed that this transcription factor activates the expression of SA-mediated defenserelated genes.These results suggest that the function of GmWRKY40 in regulating plant resistance to diverse pathogens is relatively conserved.However,due to the two duplication events that occurred during soybean evolution, there are multiple genes homologous to WRKY40 in the soybean genome.Previous studies have shown that WRKY genes may have opposite functions in plant responses to stress due to changes in their sequences [22].Therefore, whether all WRKY40 genes have similar functions in regulating plant disease resistance remains to be studied.
Investigating standing natural genetic variation is essential for understanding the evolution and selection patterns imposed during plant domestication and improvement.We detected two polymorphisms in the coding region of the GmTOC1b gene across 2479 soybean accessions.These two polymorphisms result in amino acid changes from conserved serine to a leucine (L56) or isoleucine(I473) (in W82).In Arabidopsis, the serine residue at position 175 is the most critical site affecting TOC1 phosphorylation.When this site was mutated from serine to alanine, the phosphorylation level of TOC1 significantly decreased.Transiently expression assays showed that phosphorylation of TOC1 is required to effectively repress the expression of its target genes [62].Our results show that GmTOC1bH3exhibited the strongest inhibitory activity on the GmWRKY40 promoter, whereas GmTOC1bH1showed the weakest repressive effect.This lower inhibitory activity might be influenced by the polymorphisms at amino acid residues 56 and 473, leading to a lower phosphorylation level of GmTOC1;this hypothesis needs to be tested.An investigation of the phenotypic consequences of the GmTOC1b haplotype on SMV resistance clearly showed that accessions harboring the GmTOC1bH1haplotype displayed significantly increased tolerance to SMV strain SC3 over two years.The geographical distribution of GmTOC1bH1supports the notion that this haplotype was subjected to artificial selection in the improved cultivars from the Northern and Huang-Huai regions of China.GmTOC1bH1is an outstanding haplotype that enhances resistance to SMV by weakening the inhibition conferred by GmTOC1b onto its target genes.These results provide important candidate genes for the breeding of improved soybean cultivars resistant to SMV in the future.
CRediT authorship contribution statement
Yuhang Zhang:Data curation, Investigation, Methodology,Writing – original draft.Haiping Du:Data curation, Methodology.Tiantian Zhao:Software, Data curation, Investigation, Methodology.Chunmei Liao:Investigation, Validation.Tu Feng:Investigation, Funding acquisition.Jun Qin:Investigation.Baohui Liu:Conceptualization,Supervision.Fanjiang Kong:Conceptualization,Supervision.Zhijun Che:Conceptualization,Resources.Liyu Chen:Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32001502, 32001507), the China Postdoctoral Science Foundation (2020M682655), the top ten critical priorities of Agricultural Science and Technology Innovations for the 14th Five-Year Plan of Guangdong Province (2022SDZG05), Science and Technology Innovation Team of Soybean Modern Seed Industry In Hebei Province (21326313D-4), Innovation Research Project of Coarse Cereals Specialty in Guizhou Province [2019[4012]], and the Regional First-class Discipline of Ecology in Guizhou Province(XKTJ[2020]22).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.08.001.
杂志排行
The Crop Journal的其它文章
- OsSPL10 controls trichome development by interacting with OsWOX3B at both transcription and protein levels in rice (Oryza sativa L.)
- Ectopic expression of OsNF-YA8, an endosperm-specific nuclear factor Y transcription-factor gene, causes vegetative and reproductive development defects in rice
- Mechanisms of autophagy function and regulation in plant growth,development, and response to abiotic stress
- ZmDRR206 functions in maintaining cell wall integrity during maize seedling growth and defense response to external stresses
- The plasmodesmata-associated β-1,3-glucanase gene GhPdBG regulates fiber development in cotton
- The MabHLH11 transcription factor interacting with MaMYB4 acts additively in increasing plant scopolin biosynthesis
