Makinoella parva, a new species of the genus Makinoella(Oocystaceae, Trebouxiophyceae, Chlorophyta)*
2023-12-23XudongLIUChenSUJiaFENGJunpingQiLIUFangruNANGuoxiangLIUShulianXIE
Xudong LIU, Chen SU, Jia FENG, Junping LÜ, Qi LIU, Fangru NAN, Guoxiang LIU,Shulian XIE,**
1 School of Life Science, Shanxi Key Laboratory for Research and Development of Regional Plants, Shanxi University, Taiyuan 030006, China
2 Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China
Abstract Makinoella Okada is a genus of coccoid green algae belonging to the family Oocystaceae with typical morphological characteristics.Reports to date included only one type species.Since the genus is rarely reported, there is a lack of taxonomic research.In this study, 17 strains of this genus were collected from several different places in China, and their taxonomic studies were conducted based on morphological and molecular phylogenetic analysis.The phylogenetic results based on the analysis of ITS, chloroplast genes rbcL and tufA showed that the genus was divided into two branches.One of the branches comprised newly collected algae.Compensatory base changes (CBCs) and hemi-compensatory base changes (hemi-CBCs) within the secondary structure of the ITS-2 confirmed the branches as two species.Compared with the type species of Makinoella tosaensis, the cell size of this new branch was only about 50%, and included 2–4 colonial cells.Therefore, based on the smaller cell size, simpler colony composition, independent phylogenetic position and CBCs and hemi-CBCs, we suggest that the new clade should be designated as a new taxon of Makinoella, namely Makinoella parva.Among the four molecular markers using in this study, the rbcL and newly introduced ITS are recommended for species separation,which can help further studies to revise the species and generic concepts of the family.
Keyword: coccoid green algae; Makinoella; morphology observation; Oocystaceae; phylogenetic analysis
1 INTRODUCTION
MakinoellaOkada is a genus of coccoid green algae which dispose of unique morphology.It was characterized by typical 4–16 celled colonies enclosed in mother cell wall, regularly arranged in a flat plane.Tetrads of the cell were arranged in a quadrilateral shape with subterminal connection (Hegewald et al.,1999).Samples of the genusMakinoellacan be easily recognized under microscopic observation due to the large colony size and unique cell arrangement.Recently, only one species, the type speciesM.tosaensis, was recognized in the genusMakinoella.(Okada, 1949).It was record in four countries up to now (Okada, 1949; Hegewald et al., 1999; Hindák and Hindáková, 2010; Zhang et al., 2016).
Makinoella tosaensiswas first discovered in Kochi Prefecture in 1949 (Okada, 1949), and has been reported in several other regions of Japan since then(Fukushima, 1956; Yamagishi and Akiyama, 1984).It was initially classified under the family Oocystaceae(Okada, 1949).The classification was also supported by subsequent studies (Bourrelly, 1966).However,Komárek and Fott (1983) proposed that the alga should be transferred to the family Scenedesmaceae because of the flat 4-celled morphology of the colonies.Due to the similar colony arrangement withCrucigenia Lemmermann, the genusMakinoellawas further included in the subfamily Crucigenioideae(Scenedesmaceae) (Komárek and Fott, 1983).Komárek and Fott (1983) suggested a possible classification of the genusMakinoellaunder class Xanthophyceae due to the presence of numerous chloroplasts and absence of pyrenoids.Between 1996 and 1999,Makinoellawas discovered repeatedly at Sookmyung Women’s University in Seoul, Republic of Korea,and was recorded by Hegewald et al.in 1999, which was also the first time that the algae were discovered beyond Japan (Hegewald et al., 1999).Hegewald et al.(1999).performed a detailed ultrastructural examination ofMakinoella, revealing the perpendicular arrangement of cellulose fibrils adjoining the layer of cell wall.The cell wall ultrastructure unique to the members of family Oocystaceae (Robinson and Preston, 1972; Robinson and White, 1972; Montezinos and Brown, 1978) suggested that the genusMakinoellashould be reassigned to family Oocystaceae (Hegewald et al., 1999).In addition, transmission electron microscopy clearly showed thatM.tosaensiscontains a pyrenoid surrounded by starch sheath (Hegewald et al., 1999; Schnepf and Hegewald, 2000).Subsequently,Hepperle et al.(2000) sequenced the 18S rDNA gene of an algal strain (M.tosaensisSAG28.97) isolated from Korea by Korea by Hegewald et al.(1999) and finally established the taxonomic position of the genusMakinoellavia phylogenetic analysis.Accordingly,Makinoellawas included in family Oocystaceae (Hepperle et al., 2000) with the closest relationship withOocystis solitaria.Hindák and Hindáková (2010) collected a strain ofMakinoellain the Slovak Republic, which suggested the initial expansion of the genus beyond East Asia.Recently,Zhang et al.(2016) reported two strains from the Wuhan Zoo in Hubei Province and Xishuangbanna tropical botanical garden in Yunnan Province, which was the first record ofMakinoellain China.
In this study, we isolated and cultured 17 strains of genusMakinoellafrom 10 cities of 6 provinces in China.The taxonomic studies of genusMakinoellaincluded morphological observations combined with phylogenetic analysis of multiple molecular markers.The comparison of the ITS-2 secondary structure,together with compensatory base changes (CBCs)and hemi-compensatory base changes (hemi-CBCs),was also used for species separation.The results justified the description of the novel species,M.parva,within the genus.
2 MATERIAL AND METHOD
The microscopic morphology of all algal strain samples was observed using an Olympus BX53 light microscope (Olympus Corp., Tokyo, Japan) and an Olympus DP80.The sizes of 50 cells per strain were measured using Olympus cellSens software.The mucilage envelope of the algal strain was shown by negative staining with Indian ink.
DNA was extracted using a genome extraction kit (Tiangen Biotech Co., China).Four molecular markers, the 18S rDNA, chloroplast genesrbcL andtufA, and ITS region carrying ITS1, 5,8S rRNA and ITS2, were selected.The primers are shown in Table 2.The amplified product was sent to Beijing Huada Gene Corporation for sequencing.The newly obtained sequences have been submitted to GenBank.
The sequences used for the phylogenetic analysis of 18S rDNA andrbcL were selected based on studies by Štenclová et al.(2017) and Liu et al.(2017, 2018b, 2020).In the absence oftufA and ITS molecular data associated with family Oocystaceae in GenBank, all the sequences used for the phylogenetic analysis based on these two molecular markers were newly obtained in this study.All datasets were aligned using ClustalW (Larkin et al., 2007)and edited manually using Mega 5.2.2 (Tamura et al.,2011).IQ-TREE (Nguyen et al., 2015) and MrBayes 3.2.6 (Ronquist et al., 2012) were used for Maximum Likelihood Estimation (MLE) and Bayesian inference(BI) analysis.The best-fit model was selected by ModelFinder (Kalyaanamoorthy et al., 2017) and listed as Table 3.
The ITS-2 region was detected by ITS2 Database(Schultz et al., 2006).The secondary structure of ITS-2 was constructed based on the RNA structure(Mathews et al., 2004).The hemi-CBCs and CBCs were located by program 4SALE (Seibel et al.,2006, 2008).
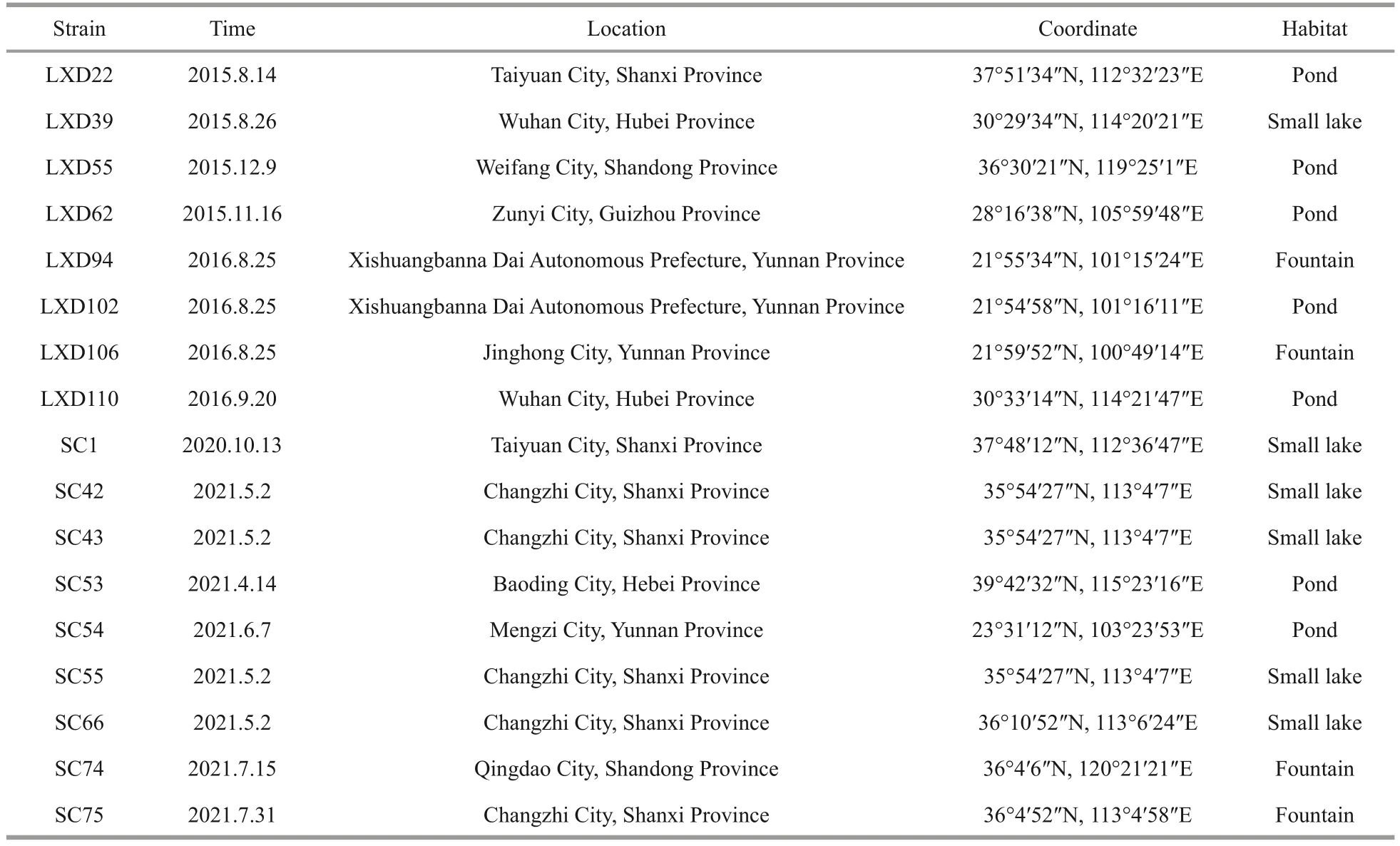
Table 1 List of algal strains sampled in this study

Table 2 Primers used in this study

Table 3 Best-fit model used in this study
3 RESULT
3.1 Morphological observation
TheMakinoella parvaconsisted of a simple colony of 2 to 4 cells, surrounded by the mother cell wall (Fig.1a & g).Occasionally, solitary cells were observed.The compound colony with multigenerations was seldom formed.The 4-celled colony was composed of two pairs of parallel and symmetrical cells (Fig.1a & e).The longitudinal axes of the cells were located at an angle (Fig.1a & i)or even almost perpendicular to each other (Fig.1c).Occasionally, the four cells were arranged in a square on the same plane (Fig.1e), and connected via subpolar regions or separated from each other following the expansion of the mother cell wall(Fig.1a & e).In a 2-celled colony, the cells formed a certain angle or even vertical distribution in different planes (Fig.1h).The cells were sometimes arranged in the same plane, parallel or non-parallel to each other(Fig.1d & g).The mother cell wall expanded with wave-like margin (Fig.1a, c, g, & h) and exhibited obvious bilateral asymmetry in a 2-celled colony(Fig.1d, g, & h).The surface of mother cell wall showed a fibrillar network structure (Fig.1d, e, & i)with radially adhering rod-like bacteria (Fig.1a & c).A thick extracolonial mucilaginous envelope was present (Fig.1d).

Table 4 Sequences used for molecular analysis of Makinoella in present study
Cells were elliptical to broadly elliptical, sometimes slightly asymmetrical (Fig.1e), and 11.2–21.4 μm long and 7.3–15.2 μm wide.The apical thickening was sometimes visible in older cells.Chloroplasts were discoid or slightly lobated, 4–8 or more, each with a pyrenoid (Fig.1b, e, & f).The pyrenoid was not always visible (Fig.1a & d).Numerous metal particles and oil droplets were present in mature cells (Fig.1c & i).Asexual reproduction involved 2 to 4 autospores.The mother cell wall expanded and then gelatinized or ruptured to release the daughter cells (Fig.1a).
TheMakinoella tosaensisshowed similar cell arrangement and colony formation withMakinoella parva.In comparison, theMakinoella tosaensishad more compound colony with 2–3 generations enclosed by old mother cell wall (Fig.1k).The colony mucilage envelope was thick (Fig.1l).The cells ofMakinoella tosaensiswere often broadly elliptical (Fig.1j), 16–25.6 μm long and 10.6–19.5 μm wide.When reproductive, the mother cell divided by a transverse wall into 2 autospores(Fig.1l).The 4 autospores were also common(Fig.1l).When the daughter cell turned matured, the similar division happened in the mother cell wall.The mother cell wall ruptured to release daughter cells.
3.2 Phylogenetic analysis
In the present study, 18S rDNA gene was sequenced in eight strains successfully (Table 4),none of which had introns.Phylogenetic analysis was performed in 106 sequences of the representatives of Oocystaceae.Ankistrodesmus fusiformis(Chlorophyta)was selected as the outgroup.Sequence alignment and editing manually yielded a dataset of 1 474 bp.Phylogenetic tree (Fig.2) of the 18S rDNA showed that the genusMakinoellawas included in the subfamily Makinelloideae and was closely related toEcballocystis hubeiensisandOocystis nephrocytioides.All the strains ofMakinoellaclustered on one clade and showed no obviously intra-genus differences.
With these words he tried to slip back into his house, but the tailor laid hold of his coat-tails, and begged so hard to be allowed to stay that the old fellow, who was by no means as cross as he appeared, was at length touched by his entreaties8, let him come in, and after giving him some food, showed him quite a nice bed in one corner of the room

Fig.1 Light microscopy of Makinoella parva and Makinoella tosaensis

Fig.2 Phylogenetic tree of Oocystaceae based on 18S rDNA
Phylogenetic analyses of therbcL cpDNA were based on selected 108 representative sequences of the family Oocystaceae including 15 newly obtained sequences of the genusMakinoellain this study.Five strains ofChlorellawere selected as the outgroup.The final dataset contained 1 037 bp ofrbcL cpDNA.Phylogenetic tree (Fig.3) showed that the genusMakinoellawas clearly divided into two main clades.The basal clade consisted of the Korean strain SAG28.97(KY710890), the Slovakian strain CCALA961(KY710879) and our strains LXD55, LXD62, SC1,and SC74.The algal strains LXD22, LXD39, LXD102,LXD106, LXD110, SC42, SC43, SC53, SC55, SC66,and SC75 clustered into the derivative clade with full support.
Nine sequences were used for phylogenetic analysis oftufA of the subfamily Makinelloideae, including 7 newly sequencedMakinoellastrains and the other two strains,Ecballocystissp.LXD88 andReticulocystis yunnanenseLXD107, as the outgroup.The sequences were aligned and manually edited in a matrix containing a total of 873 bp of base sites.The phylogenetic tree(Fig.4) also showed that algal strains LXD22, LXD39,LXD102, LXD106, and LXD110 clustered into one clade, clearly separated from algal strains LXD55 and LXD62 with high support values (100/0.9).
A total of 16 ITS sequences belonging to the genusMakinoellawere newly obtained.Sequences of two other representatives of subfamily Makinoelloideae,Ecballocystissp.LXD88 andReticulocystis yunnanenseLXD107, were used as outgroups.The final matrix contained a total of 620 bp of base sites.The ITS phylogenetic tree (Fig.5) constructed using MLE and BI analysis showed that the genusMakinoellaconsisted of two main clades, with strains LXD22, LXD39,LXD94, LXD102, LXD106, LXD110, SC42, SC43,SC53, and SC55 clearly separated from the clade formed by the algal strains LXD55, LXD62, SC54,and SC74, which was also strongly supported statistically (92/0.94).
The consensus ITS2 secondary structure model ofMakinoellais shown in Fig.6.Based on the comparison of the ITS2 secondary structure, we found 2 hemi-CBCs in helix III and 1 hemi-CBC in helix II between the two species.
4 DISCUSSION
Makinoellais a rare genus of green algae reported only six times worldwide (Okada, 1949;Fukushima, 1956; Yamagishi and Akiyama, 1984;Hegewald et al., 1999; Hindák and Hindáková,2010; Zhang et al., 2016).In this study, 17 isolated and cultured algal strains (Table 1) suggest that the genusMakinoellahas a wide geographical distribution in China.Our findings together with previous studies indicate that theMakinoellaspecies inhabit still waters with small artificial substrates, such as pond, fountains,and small lakes (Okada, 1949; Hegewald et al., 1999;Hindák and Hindáková, 2010; Zhang et al., 2016).
This study entailed a detailed taxonomical investigation of the genusMakinoella, based on morphological observations combined with molecular phylogenetic analysis.The evolutionary relationships within the genus were analyzed using ITS andtufA cpDNA molecular markers for the first time in the family Oocystaceae.Phylogenetic analyses based on ITS,rbcL cpDNA, andtufA cpDNA showed that the genus was clearly divided into two branches with high statistical support, which implied the evolution of at least two distinct taxonomic units in this genus(Figs.3–5).The CBCs content in the ITS-2 between the branches further supported the separation as two species.Nevertheless, the molecular sequences of the type species have yet to be identified.Therefore, we were unable to classify accurately these two species based on molecular phylogenetic analysis alone.
Morphological comparisons of the two clades ofMakinoellastrains mentioned above were further conducted.The basal clade consisted of the Korean strain SAG28.97 (Hegewald et al., 1999), the Slovakian strain CCALA961 (Hindák and Hindáková, 2010)and the strains LXD55, LXD62, SC54, and SC74 obtained in this study (Fig.3) with similar morphology.The morphological characteristics of cells in this clade were similar to the type species, and some of the differences were attributed to not enough detailed observation, which led to wrong original description,and all the strains contained pyrenoids (Okada,1949).In the family Oocystaceae, the pyrenoid were not always visible, especially when the cells carried a high number of starch grains, oil droplets and other inclusions (Řeháková, 1969), which were typical ofM.tosaensis(Hegewald et al., 1999).Therefore,the species of this clade was designated asM.tosaensis, based on the identification of Hegewald et al.(1999) and Hindák and Hindáková (2010).

Fig.3 Phylogenetic tree of Oocystaceae based on rbcL cpDNA
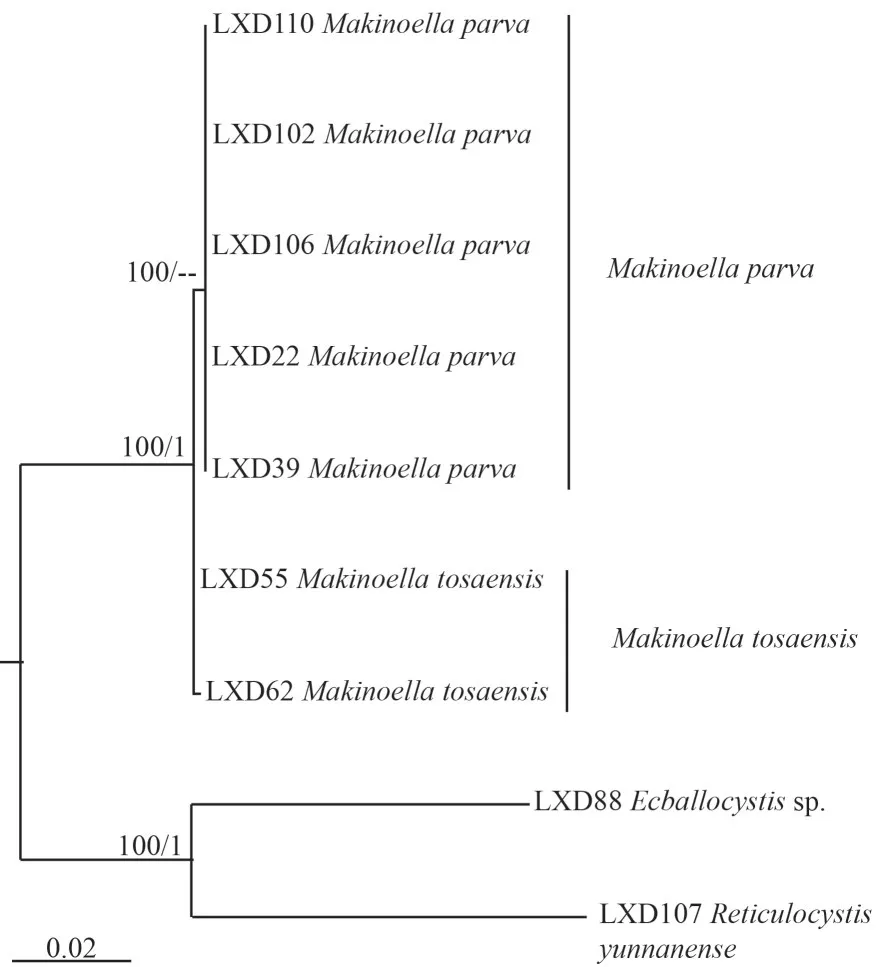
Fig.4 Phylogenetic tree of Makinoella based on tufA cpDNA
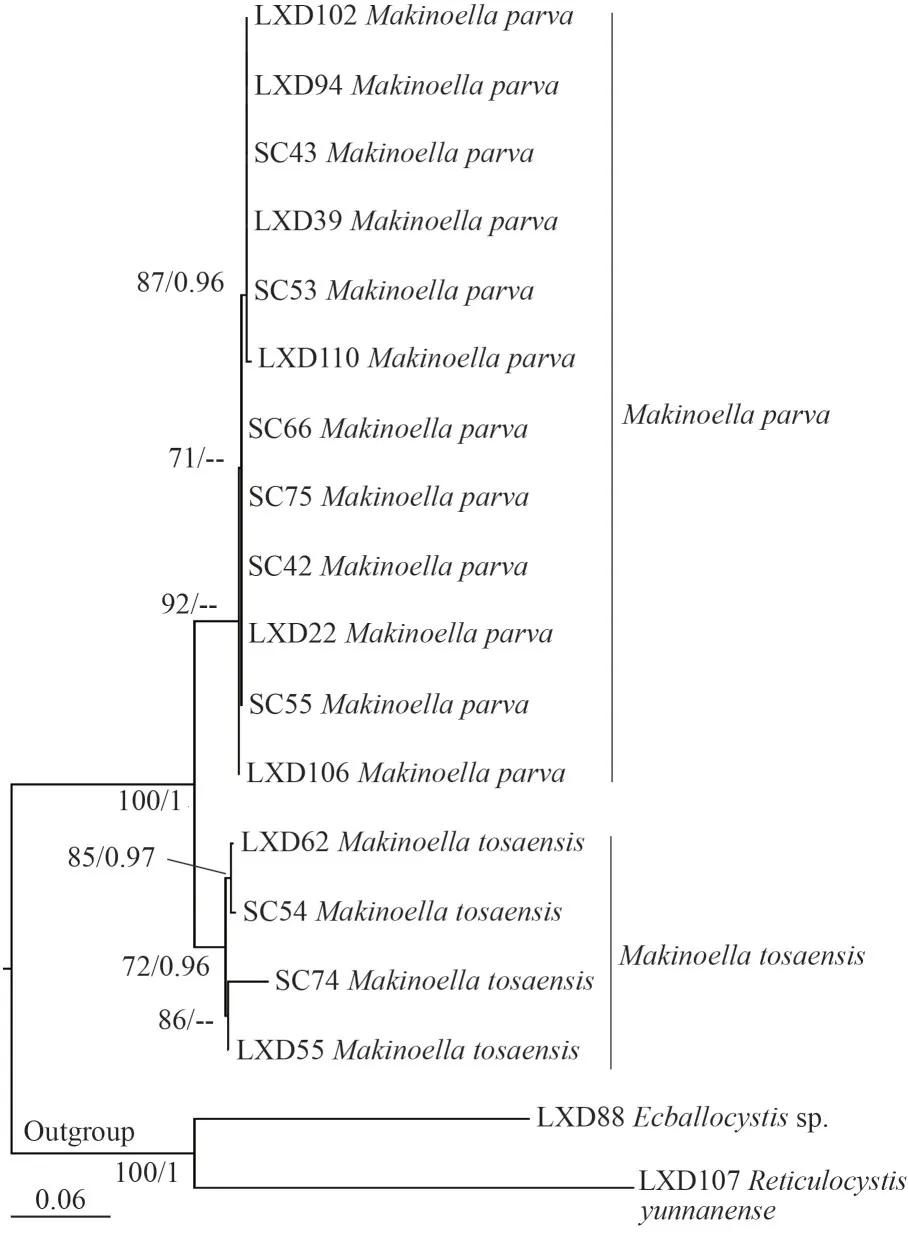
Fig.5 Phylogenetic tree of Makinoella based on ITS
The other clades included newly identified algal strains LXD22, LXD39, LXD94, LXD102, LXD106,LXD110, SC42, SC43, SC53, SC55, SC66, and SC75 (Figs.3–5), which were significantly smaller in size compared with the type species ofMakinoella.The cell size of the type species ((22–30) μm×(16–18) μm) was approximately twice that of the new clades ((11.2–21.4) μm× (7.3–15.2) μm), as shown in Table 5.Cell size was one of the important taxonomic characteristics in the family Oocystaceae(Řeháková, 1969; Komárek and Fott, 1983; Stoyneva et al., 2007), and many species and varieties have been described accordingly, such asOocystis parva,Quadricoccopsis parva, andEcballocystis pulvinatavar.minor(West and West, 1898; Iyengar, 1932; Liu et al., 2018b).Our results show that the cell size was relatively constant among different strains of the same species (wild or cultured), which was also supported by previous studies ofMakinoellareported by Hegewald et al.(1999).Therefore, the cell size was recognized as an effective morphological feature for the classification of the genusMakinoella.Furthermore, the composition of the new clade revealed relatively simpler colonies composed of 2–4 cells, and rarely included multiple generations.In comparison, the type species consisted of 4–16 cells forming simple or compound colonies.Colony composition was also one of the distinguishing characteristics of the family Oocystaceae on species and generic levels (Komárek and Fott, 1983; Liu et al.,2018b).Based on cell size and colony characteristics,the genusQuadricoccopsiswas subdivided into three species:Q.simplex,Q.parva, andQ.glomerata(Liu et al., 2018b).Therefore, based on the smaller cell size, simpler colony composition and independent phylogenetic position, we described the new clade as a new species,M.parva, within the genusMakinoella.
Among the four molecular markers used in this study, 18S rDNA was the most widely used in taxonomical studies of small coccoid green algae.However, the increased data revealed lower variation and phylogenetic signals of 18S rDNA, which interfered with species separation (Bock et al., 2011; Wang etal., 2019).In genusMakinoella, the 18S rDNA also failed to distinguish the different species based on our results.Therefore, distinguishing different genera in Oocystaceae was more appropriate, similar to Scenedesmaceae (Sciuto et al., 2015).TherbcL gene has been associated with Oocystaceae (Liu et al., 2017,2018a, b, 2020, 2021; Štenclová et al., 2017) and other taxa in Trebouxiophyceae (Song et al., 2015;Sanders et al., 2016).It showed high efficiency for interspecific separation.By contrast, thetufA gene has been recommended for easier amplification and sequencing than therbcL in the taxonomic study of Chlorophyceae (Vieira et al., 2016), especially in Scenedesmaceae (Sciuto et al., 2015; Wang et al.,2019, 2020).However, in Oocystaceae, the situation was different.Although both are effective for interspecific discrimination of genusMakinoella, therbcL showed relatively higher amplification efficiency than thetufA.In addition, therbcL sequences were abundant in the comprehensive analysis by Štenclová et al.(2017), thereby supportingrbcL.In contrast to the two chloroplast genes mentioned above, the ITS molecular markers showed the highest variability.They have been widely used for species separation in Trebouxiophyceae (Krienitz et al., 2004; Bock et al., 2013; Malavasi et al., 2016;Song et al., 2018), especially in Chlorellaceae (Luo et al., 2006, 2010; Krienitz et al., 2010, 2012; Bock et al., 2011).In Oocystaceae, it was also very efficient for the interspecific or intraspecific classification,which was supported by the CBCs and hemi-CBCs results.This study fully supported the application of ITS in the taxonomic study of Oocystaceae.Additional ITS data will be significant for the discovery of species diversity in this family.
Taxonomic assessment
Makinoella parvaLIU, SU, LIU et XIE sp.nov.(Fig.1)
Diagnosis: Colony free-floating, 2–4 cells surrounded by the mother cell wall; cells often not on the same plane or almost perpendicular to each other; mother cell wall wave-like and often with bilateral asymmetry; colonies with thick mucilaginous envelope; cells elliptical to broadly elliptical, 11.2–21.4 μm long and 7.3–15.2 μm wide; chloroplasts 4–8 or more per cell, discoid or small lamellate,each with a pyrenoid; propagation asexually by 2 or 4 autospores.Species differ fromM.tosaensisdue to smaller cell size, simpler colony composition and the order of nucleotides inrbcL cpDNA,tufA cpDNA, and ITS.
Type: Baoding City, Hebei Province, China: sample collected in a pond, 39°42′32″N, 115°23′16″E, 14/4/2021, Chen SU (Holotype SXU-SC53).Algae partially illustrated here in light microscopy (Fig.1).
Reference strain: the living strain was deposited in Freshwater Algae Culture Collection at Institute of Hydrobiology, Wuhan, China as No.FACHB-3564.
Habitat: pond; planktonic; water temperature=17.6 °C; pH=8.41.
Etymology: the species is named for the smaller cell size compared with the other species of this genus.
5 CONCLUSION
The study establishes a new species of the genusMakinoella,M.parva, based on smaller cell volume and simple colony composition and independent phylogenetic position.TherbcL cpDNA and newly used ITS molecular makers showed higher efficiency for interspecific discriminations, and therefore recommended for future taxonomic studies of Oocystaceae.
6 DATA AVAILABILITY STATEMENT
The authors declare that the datasets in this study are available on reasonable request from the corresponding author.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Trends of carbon and nutrient accumulation through time in the Andong salt marsh, Hangzhou Bay, China*
- Physical processes determining the distribution patterns of Nemopilema nomurai in the East China Sea*
- Comparison in structure and predicted function of epiphytic bacteria on Neopyropia yezoensis and Neopyropia katadae*
- Interaction between macroalgae and microplastics: Caulerpa lentillifera and Gracilaria tenuistipitata as microplastic bio-elimination vectors*
- Lake regime shift from submerged macrophyte to phytoplankton affected phosphorus speciation in sediment and eutrophic state in Caohai Lake, Guizhou, China*
- Temporal characteristics of algae-denitrifying bacteria co-occurrence patterns and denitrifier assembly in epiphytic biofilms on submerged macrophytes in Caohai Lake, SW China*
