Genome-wide characterization of mapk gene family in black rockfish Sebastes schlegelii and their expression patterns against Edwardsiella piscicida infection*
2023-12-23XiaoyanZHANGYuanTIANHaohuiYUMinCAOChaoLI
Xiaoyan ZHANG, Yuan TIAN, Haohui YU, Min CAO, Chao LI,**
1 School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao 266109, China
2 Key Laboratory of Mariculture (Ocean University of China), Ministry of Education, Ocean University of China, Qingdao 266003, China
Abstract Mitogen-activated protein kinases (MAPKs) play pivotal roles in response to environmental stresses and bacterial infections.Compared with those in the higher vertebrates, studies of mapk gene family are still limited in teleost.Identification, characterization, classification, and expression profiling of totally 15 mapk genes in black rockfish (Sebastes schlegelii) were conducted.Phylogenetic relationships show that these mapk genes could be divided into extracellular signal-regulated kinase(ERK), c-Jun N-terminal kinase (JNK), and p38 sub-families.In addition, gene structures, syntenic analysis, and selective pressure analysis are performed to confirm their annotations.Results of selective pressure analysis indicate that mapk1, mapk3, mapk7, mapk10, mapk11, and mapk12 underwent significantly-positive selections, while the others genes such as mapk4, mapk6, mapk15, mapk8a, mapk8b,mapk9, mapk13, mapk14a, and mapk14b were under purifying selections.Moreover, results of qRT-PCR indicate that mapk genes in 8 healthy tissues displayed different expression patterns.The expression patterns of several mapk genes including mapk12, mapk13, mapk14a, mapk14b, and mapk15 were significantly changed in mucosal tissues after Edwardsiella piscicida infection.This study demonstrates that mapk genes in black rockfish play vital prevention roles against bacterial infection, which not only helps us understand the structure and function of mapk genes in black rockfish, but also provides a reference to understand the role of mapk genes in teleost immune responses.
Keyword: Sebastes schlegelii; mitogen activated protein kinases (mapk); Edwardsiella piscicida;expression profiles
1 INTRODUCTION
Mitogen-activated protein kinases (MAPKs) are class of serine/threonine protein kinases that activated by extracellular stimuli through the MAPK cascade that widely presented in most cells (Seger and Krebs, 1995; Cowan and Storey, 2003; Roux and Blenis, 2004; Sun et al., 2015).Usually,MAPKs mainly contain extracellular signalregulated kinases (ERK), c-Jun amino-terminal kinases (JNK), and p38-subfamily, those were classified according to the differences in dualphosphorylation sites (Krens et al., 2006).Being well known, phosphorylated MAPKs can transduce extracellular signals into intracellular responses,activate downstream molecules, and regulate the expressions of target genes in various biological processes such as growth, development, apoptosis,and responses to various environmental stresses and bacterial infections (Garrington and Johnson, 1999;Cowan and Storey, 2003; Kyriakis and Avruch,2012).Accordingly, numerous literatures focused on the structures and functions ofmapkgenes in both mammals and teleost.For instance, it was reported that ERK could be activated by oxidant stress in kidney of mouse (Mus musculus) (Arany et al.,2005); ERK2, ERK3, and p38A were expressed at constant levels during zebrafish embryogenesis as proved by semi-quantitative reverse transcriptase-PCR analysis and in-situ hybridization (Krens et al.,2006).Tian et al.(2019) identified 14mapkgenes in spotted sea bass (Lateolabraxmaculatus), and speculated thatmapkgenes such as JNK (JNK1,JNK2) and p38 (p38β,p38a, andp38b) were involved in the response to salinity and hypoxia challenges.
Additionally, many studies have clarified the functions ofmapkgenes in immune response ranging from mammals to teleost.In mammalian species,mapkgenes were believed closely associated with the innate immunity and adaptive immunity(Dong et al., 2002).For teleost, the conservedmapkgenes were considered as critical mediators of innate immune system for regulating the expression of cytokines to defend against various types of pathogen infection (Zhu et al., 2014; Jia et al., 2015;Umasuthan et al., 2015; Sun et al., 2020).For example, the injection ofAeromonas hydrophilaand muramyl dipeptide in the intestine of grass carp(Ctenopharyngodonidella) can induce the expression ofCip38andCip38β, which also can influence the expression of inflammatory cytokines and antimicrobial peptides.This study proved the crucial roles of p38genes in bacteria-induced intestinal inflammation (Sun et al., 2020).Moreover,we found that ERK genes elicit a strong immune response to the stimulation of polyI꞉C and flagellin in large yellow croaker (Larimichthys crocea) (Jia et al., 2015).Similar findings were also observed in other teleosts, such as Atlantic salmon (Salmo salar)(Hansen and Jørgensen, 2007), grouper rock bream(Oplegnathus fasciatus) (Umasuthan et al., 2015),orange-spotted grouper (Epinephelus coioides) (Sun et al., 2017), and blunt snout bream (Megalobrama amblycephala) (Zhang et al., 2019).These published studies demonstrated thatmapkgenes participated in the immune response and the regulation of defensing activities in teleost (Menon et al., 2017;He et al., 2018; Zhang et al., 2019; Cheng et al.,2020; Wei et al., 2020).
Black rockfish(Sebastes schlegelii) is an economically important mariculture species in the northern China.Expansion of aquaculture industry due to market demand has resulted in frequent outbreaks of various bacterial diseases, causing serious economic losses to the farming industry(Kitamura et al., 2006; Cao et al., 2020).Studying the antibacterial mechanism of black rockfish is beneficial to disease prevention.To investigate the potential roles ofmapkgenes inblack rockfish upon infection with pathogens, a systematically identification and characterization ofmapkgenes based on genome and transcriptomewere performed in this study.Their characteristics, phylogenetic relationships, and selective pressures were analyzed.Moreover, the expression patterns of 15mapkgenes in healthy tissues of black rockfish were detected by qRT-PCR, as well as their response expressions followingEdwardsiella piscicidainfection, aiming to provide a fundamental resource to understand the potential immune functions ofmapkgenes of black rockfish.
2 MATERIAL AND METHOD
2.1 Identification of mapk genes of black rockfish
To identifymapkgenes of black rockfish, we firstly downloaded several sequences of MAPKs from available species including human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), and several teleost from the NCBI database.These genes were chosen as queries for searching the candidates ofmapkgenes using TBLASTN.Thus, the candidatemapkgenes from black rockfish were obtained against available whole genome sequence and transcriptome sequences with E-value less than 1e-10.Meanwhile, we confirmed these genes by predicting their open reading frame(ORFs) using the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and their NR annotations.
2.2 Phylogenetic relationships and collinearity analysis
The amino acid sequences ofmapkgenes from several representative vertebrates including human,mouse, chicken, cow (Bos taurus), frog (Ceratophrys cornuta), zebrafish, Nile tilapia (Oreochromis niloticus), Atlantic salmon, large yellow croaker,torafugu (Takifugu rubripes), and Japanese medaka(Oryzias latipes) that were downloaded from the NCBI database, and those that were identified in this study were aligned, to clarify the evolutionary relationships ofmapkgenes in black rockfish.In detail, software MUSCLE (MUltiple Sequence Comparison by Log-Expectation) was employed to perform multiple amino acid sequences (Edgar,2004).Subsequently, software MEGA 7 was used to construct the neighbor-joining (NJ) phylogenetic tree with 1 000 bootstraps (Kumar et al., 2016).Finally, the iTOL (the Interactive Tree of Life) was used to display and embellish the tree (http://itol.embl.de/).
In addition, collinearity analysis was performed to confirm the accuracy of identification and annotation of thesemapkgenes, especially for the duplicated ones.The neighboring genes aroundmapkgenes in black rockfish were identified from its genome.The genomic information of zebrafish was obtained from NCBI, Genomicus, and Ensembl databases.Meanwhile, annotation and their chromosome locations of thesemapkgenes were obtained from the genome of black rockfish.At last,software TBtools was used to present the chromosomal locations ofmapkgenes (Chen et al.,2020).
2.3 Gene structure, domain, and motif analysis
The information about the numbers of exonintron, functional domains and motifs permapkgene of black rockfish were presented with TBtools(Chen et al., 2020).Molecular weights and theoretical isoelectric point (pI) of these proteins was calculated using ProtParam.Information about gene structures was extracted from the Nr annotation results.In addition, SMRT database was utilized to predict the architectures of the typical domain ofthesemapkgenes.The conserved protein motifs of eachmapkgenewere identified with MEME (Bailey et al., 2009), then were annotated by InterProScan (Mulder and Apweiler, 2007).Additionally, we compared the copy numbers ofmapksin vertebrates including black rockfish.
2.4 Selective pressure analysis
To study the selective pressures on thesemapkgenes during evolution, different amino acid sequences of eachmapkgene from human, mouse,chicken, cow, frog, Atlantic salmon, zebrafish,torafugu, Japanese medaka, and black rockfish were firstly aligned using software MUSCLE (Edgar,2004).Subsequently, we used the PAL2NAL program to perform the codon alignments in coding DNA in the estimation of type and ratio of nucleotide substitutions.Meanwhile, the NJ tree of eachmapkgene was constructed according to the aligned amino acid sequences using MEGA 7 software with the JTT model.Thus, dN/dS with branch-site model were compared to generate the selective pressure of eachmapkgene among these species using PAML program (Yang, 2007).P<0.05 was chosen to assess the significance of the alternative hypothesis.
2.5 Sample collection from the healthy tissues of S. schlegelii
All procedures involving the handling and treatment of fish in this study were approved by the Institutional Animal Care and Use Committee,Qingdao Agricultural University.The individuals of black rockfish (body length 15±0.55 cm and body weight 110±5 g) were sampled from Weihai,Shandong, China, and cultured in laboratory for one week before experiment.During the period of experiment, the environmental conditions were kept relatively constant.To understand the expression levels of thesemapkgenes in healthy tissues ofS.schlegelii, eight tissues (brain, gill, muscle,kidney, liver, spleen, intestine, and skin) were collected to detect the expression patterns of 15mapkgenes.During sampling collection,individuals were anesthetized with MS-222 (Sigma Aldrich Co., St.Louis, MO, USA).After that, eight tissues were collected separately from 9 fish for RNA extraction.Tissues from three fish individuals were mixed as one sample.
2.6 Bacterial infection experiment
For exploring the expression profiles ofmapkgenes in black rockfish followingE.piscicidainfection, this bacterium was firstly isolated from diseased fish.Then,E.piscicidawas cultured in LB medium with 180 r/min at 28 °C.The concentration ofE.piscicidaused in the experiment was calculated by counting colony forming unit (CFU)onto plates.Finally, the individuals immersed 4 h inE.piscicidawith a final concentration of 1×107CFU/mL.In contrast, individuals in seawater were collected as control.Subsequently, skin, gill,and intestine were collected at 0 h, 2 h, 6 h, 12 h,24 h, and 48 h time points.Then, the collected samples were stored in liquid nitrogen for RNA extraction.
2.7 RNA extraction, cDNA synthesis, and qRTPCR experiment
Trizol method (Invitrogen, Carlsbad, CA, USA)was used to extract RNA from different tissues,including 8 healthy tissues, mucosal tissues followingE.piscicidainfection.After extraction,RNase-Free DNase I (TIANGEN, Beijing, China)was added into the extracted RNA to digest the contaminating DNA according to the manufacturer’s instructions.Afterthat, 1% agarose gels and Biodropsis BD-1000 nucleic acid analyzer(OSTC, Beijing) were used to detected the integrity and concentration of the extracted RNA,respectively.RNA with A260/280 and A260/230 over 1.8 were used in this study.Then, 1-μg RNA was reverse transcribed into cDNA synthesis using PrimeScriptTMRT reagent Kit (TaKaRa, Japan)following the manufacturer’s instructions.Subsequently, qRT-PCR was used to detect the expression levels ofmapkgenes in 8 healthy tissues and 3 mucosal tissues (skin, gill, and intestine)followingE.piscicidainfection in black rockfish.TheEF1Awas selected as a house-keeping gene for normalization (Ma et al., 2013).The primers used in this study were designed using software Primer 5 and primers are listed in Supplementary Table S1.Before experiment, the amplification efficiency of each primer was confirmed.In detail, the target amplicon of each primer was collected from gel extraction.Then, the purified PCR amplicon was cloned into pMD19-T vectorand transformed intoEscherichia coli, followed by plasmid extraction.Then, the 10-fold serial diluted standard plasmid DNAs were used as templates to generate standard curve and calculate the amplification efficiency of each primer.Those with amplification efficiency from 90% to 105% were used for qRT-PCR experiment.After validation, qRT-PCR experiment was performed on CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA).Three biological replicates and technical replicates for each sample were performed.Finally, the relative expression levels were calculated using the 2-ΔΔCtmethod (Livak and Schmittgen, 2001).The relative expression levels of the detected genes in qRT-PCR were presented as mean±standard deviation (SD).Software SPSS Statistical v17.0 was used to perform statistical analysis.P<0.05 presented significant differences between samples.
3 RESULT
3.1 Characterization of mapk genes in black rockfish
Totally, 15mapkgenes were identified in black rockfish based on available databases.The detailed characteristics of thesemapkgenes are summarized in Table 1.Based on the dual-phosphorylation sites,thesemapkgenes can be divided into three subfamilies:mapk1,mapk3,mapk4,mapk6,mapk7,andmapk15belongingto ERK subfamily;mapk8a,mapk8b,mapk9,andmapk10belonging to JNK subfamily; andmapk11,mapk12,mapk13,mapk14a,andmapk14bbelonging to p38 subfamily.In detail,ERK, JNK, and p38 harbored the typical amino-acid residues Thr-Glu-Tyr, Thr-Pro-Tyr, and Thr-Gly-Tyr,respectively.Additionally, the lengths of the amino acids of thesemapkgeneswere ranged from 345 to 1 123 bp, the values of molecular weight (MW)varied from 39.75 to 122.75 kDa, and the pIs varied from 4.93 to 9.03.
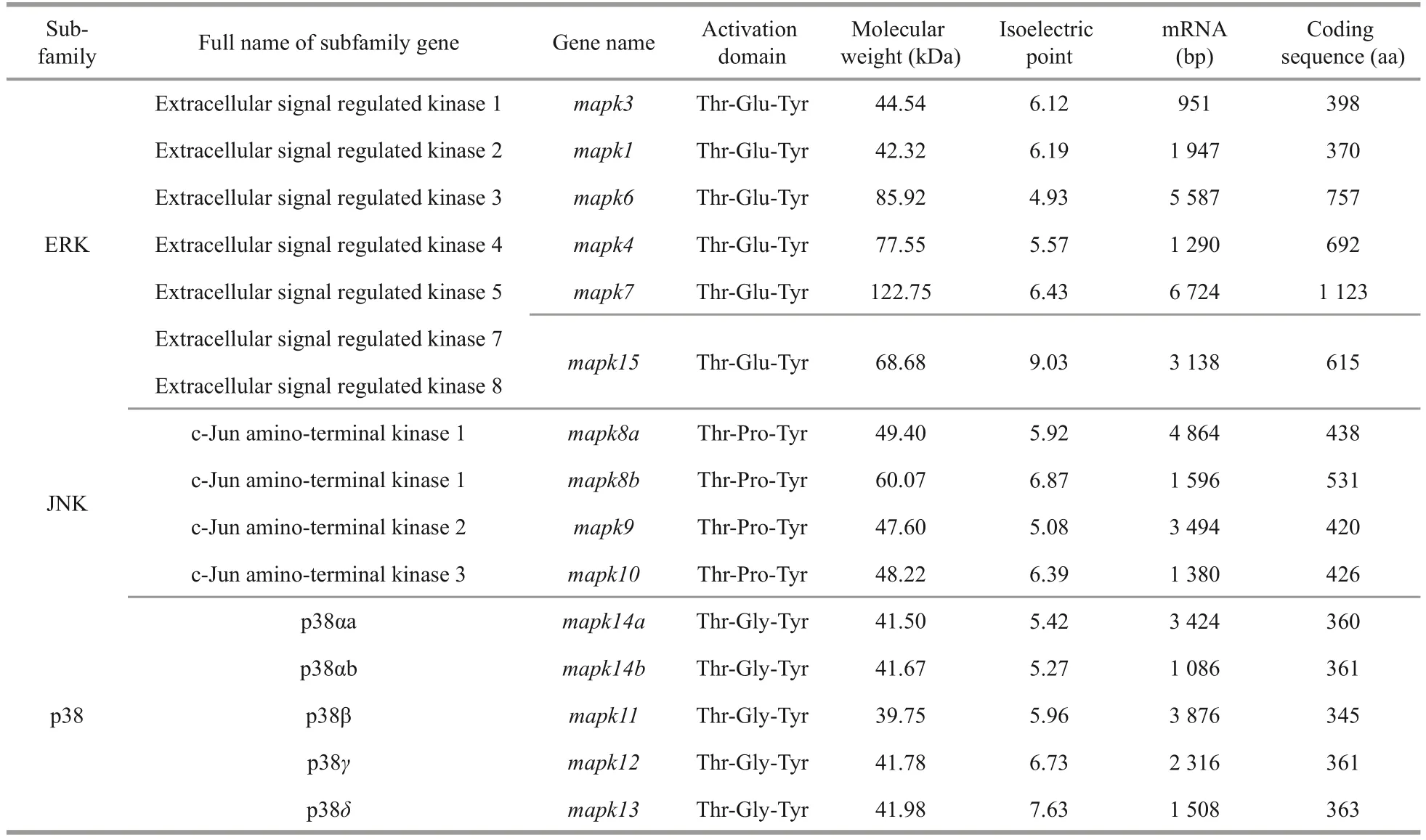
Table 1 Identification and characterization of 15 mapk genes in black rockfish
Copy numbers ofmapkgenes were examined in several high vertebrates, teleosts including black rockfish (Fig.1).Overall, the copy numbers were relatively conserved in the selected species, ranging from 10 to 15.For instance, 13, 13, 10, 12–15 members ofmapkgenes have been identified in human, mouse, chicken, and teleosts, respectively.Mostmapkgenes were present with a single copy,while duplicatedmapk8andmapk14genes were detected in several teleost, including zebrafish, large yellow croaker, spotted sea bass, turbot, and black rockfish.Thus, we speculated that the duplication events ofmapkgenes seem to be teleost-specific.
3.2 Phylogenetic relationships and syntenic analysis of mapk genes in black rockfish
For phylogenetic tree construction, amino acid sequences ofmapkgenes from black rockfish and several species were selected.Results show that thesemapkgenes are clustered with respective counterparts as expected and clearly divided into three clusters containing ERK, JNK, and p38, which were consistent with the classification ofmapkfamilies (Fig.2).The results show the accuracy of the annotation and classification ofmapkgenes in black rockfish.
Syntenic analysis was performed for further confirmation ofmapkannotation, especially for those with duplicated copies (Fig.3; Supplementary Table S2).It is obvious that the syntenic blocks surrounding the testedmapk8a,mapk8b,mapk14a,andmapk14bare similar between zebrafish and black rockfish.This result also confirms the identification and annotation of duplicatedmapkgenes in black rockfish.Moreover, in zebrafish and black rockfish, there were several paralogous genes located in the neighboring regions of duplicatedmapk8andmapk14, such asvstm4a,vstm4b,gdf10a,gdf10b,antxrld,antxrldc,srpk1a,andsrpk1b.It demonstrates that these duplicatedmapkgenes could be derived from the events of wholegenome duplication.
3.3 Chromosomal distribution, gene structure,functional domain and conserved motif analysis of mapk genes

Fig.1 Copy numbers of mapk genes in 13 representative vertebrates
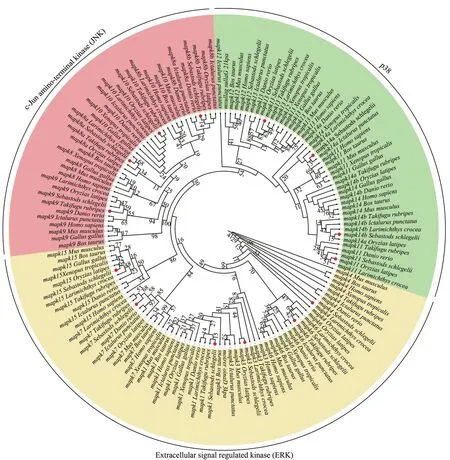
Fig.2 Phylogenetic analysis of mapk genes from the representative vertebrates
Chromosomal distribution analysis show that these 15mapkgenes were unevenly distributed on 11 chromosomes (Chrs) of black rockfish.For example, Chr1, Chr2, Chr5, Chr6, Chr7, Chr8,Chr12, Chr14, Chr16, Chr18, and Chr12.Chr1,Chr6, Chr7, and Chr18 harbored 2mapkgenes,while the other 7mapkgenes were located on different chromosomes separately (Fig.4).Meanwhile,the gene structures such as organization of exonintron in thesemapkgenes were examined in black rockfish.As shown in Fig.5a, the exon numbers of 15mapkgenes varied from 5 to 13, and gene members from same subfamily shared similar exon number.Specifically, most genes from ERK subfamily possessed 5–7 exons, except formapk15with 13 exons.In JNK subfamily, gene structures ofmapk8a,mapk8b,mapk9, andmapk10, were all comprised of 10 exons.In addition, 11 exons were observed in four p38 genes (mapk11,mapk12,mapk14a, andmapk14b), while the other p38 gene,mapk13, contained only 5 exons.The diverse exonintron structures ofmapkgenes from different subfamilies may be related to their distinct biological functions.The S_TKc domains were detected on each of MAPK proteins in black rockfish (Fig.5b).A majority of MAPK proteins contained low complexity regions, while coiled coil region was present in mapk7 only.Motifs were identified to further interpret the structural diversity ofmapkgenes in black rockfish.It is obvious that thesemapkgenes displayed similar motif arrangements (Fig.5c).In detail, motifs 1–6 and 9–10 distributed in all the identifiedmapkgenes of black rockfish, except formapk4,mapk6, andmapk11.Motif 7 was found in themapkmembers of JNK subfamily only.However, motif 8 was present in ERK and p38 subfamilies mainly.These specific motifs may be related to the functions of differentmapksubfamily genes in black rockfish.

Fig.3 Syntenic analysis of duplicated mapk8 and mapk14 genes in zebrafish and black rockfish
3.4 Selective pressure analysis of mapk genes
Eachmapkgene of the representative species was selected to construct the corresponding phylogenetic tree.The clades of black rockfish were labeled as foreground branches for the estimation of selective pressure using branch-site model (Fig.6;Supplementary Fig.S1).Results clearly show that alternative hypothesis fit significantly better than null hypothesis inmapk1,mapk3,mapk7,mapk10,mapk11, andmapk12(Table 2), demonstrating that thesemapkgenes underwent positive selection in black rockfish.Of these selected genes, the posterior probability of 10 amino acid sites was found greater than 0.95, which underwent significantly positive selection in black rockfish.However, othermapkgenes seem to be relatively conserved during the evolution.
3.5 Expression profiles of 15 mapk genes in healthy tissues
To explore the expression patterns ofmapkgenes in black rockfish under normal physiological conditions, qRT-PCR were conducted to determine the expression levels of 15mapksin 8 different tissues.As shown in Fig.7,mapkgenes displayed distinct expression levels in different tissues.Notably, mostmapkgenes witnessed relatively high expression levels in brain, especially formapk8aandmapk13.In skin,mapk1,mapk13,mapk6, andmapk15genes showhigher expression abundance.Among them,mapk1andmapk15show high expression levels in muscle, kidney, and spleen.In liver,mapk14ashow the highest expression level.Additionally,mapk12andmapk14bwere expressed in intestine mainly, whilemapk1,mapk13, andmapk14ain gill kept at higher expression levels.
3.6 Mapk genes were reduced by E. piscicida infection in black rockfish
In order to detect the response roles ofmapkgenes followingE.piscicidainfection, qRT-PCR was performed to determine the expression patterns of severalmapkgenes with abundant expression levels in intestine, skin, and gill at different time points (0 h, 2 h, 6 h, 12 h, 24 h, and 48 h) afterE.piscicidainfection.The results indicate that these selectedmapkgenes can be induced when black rockfish was infected.In intestine,mapk12,mapk14b, andmapk15significantly up-regulated at 6 h and 12 h after infection (Fig.8).Similarly,mapk1witnessed significant up-regulation at 24 h,andmapk13significantly up-regulated at 6 h, 12 h,and 48 h after infection in skin.In contrast, the expression level ofmapk14ain skin was significantly down-regulated at all detected time points.Dramatic up-regulation ofmapk15in skin was observed at 2 h, while down-regulation appeared at 6 h and 24 h.In gill,mapk1,mapk13,mapk14a, andmapk15were all significantly upregulated at 6 h, reached the peak at 24 h and then recovered to normal levels.Notably,mapk14adramatically increased with more than 17.01 folds at the peak (12 h), demonstrating that it plays an important role in the response toE.piscicidainfection in gill of black rockfish.Additionally,mapk15exhibited diverse expression patterns among intestine, skin, and gill, suggesting thatmapk15may play diverse roles in different mucosal tissues in black rockfish.
4 DISCUSSION
It is well known that MAPKs play essential roles in various biological processes by regulating related signal pathways (Seger and Krebs, 1995; Westfall et al., 2008; Pathak et al., 2013; Danquah et al., 2014).So far, arduous efforts have been made on the identification of gene families based on genome and transcriptome database in numerous plants and high vertebrates (Cargnello and Roux, 2011; Lu et al.,2015; Wang et al., 2018).However, studies related tomapkgene family remained limited in teleost.This study utilized bioinformatic and experimental approaches to identifymapkgenes in black rockfish,investigated their evolutionary relationships, and elucidated their roles in response to pathogens infection.
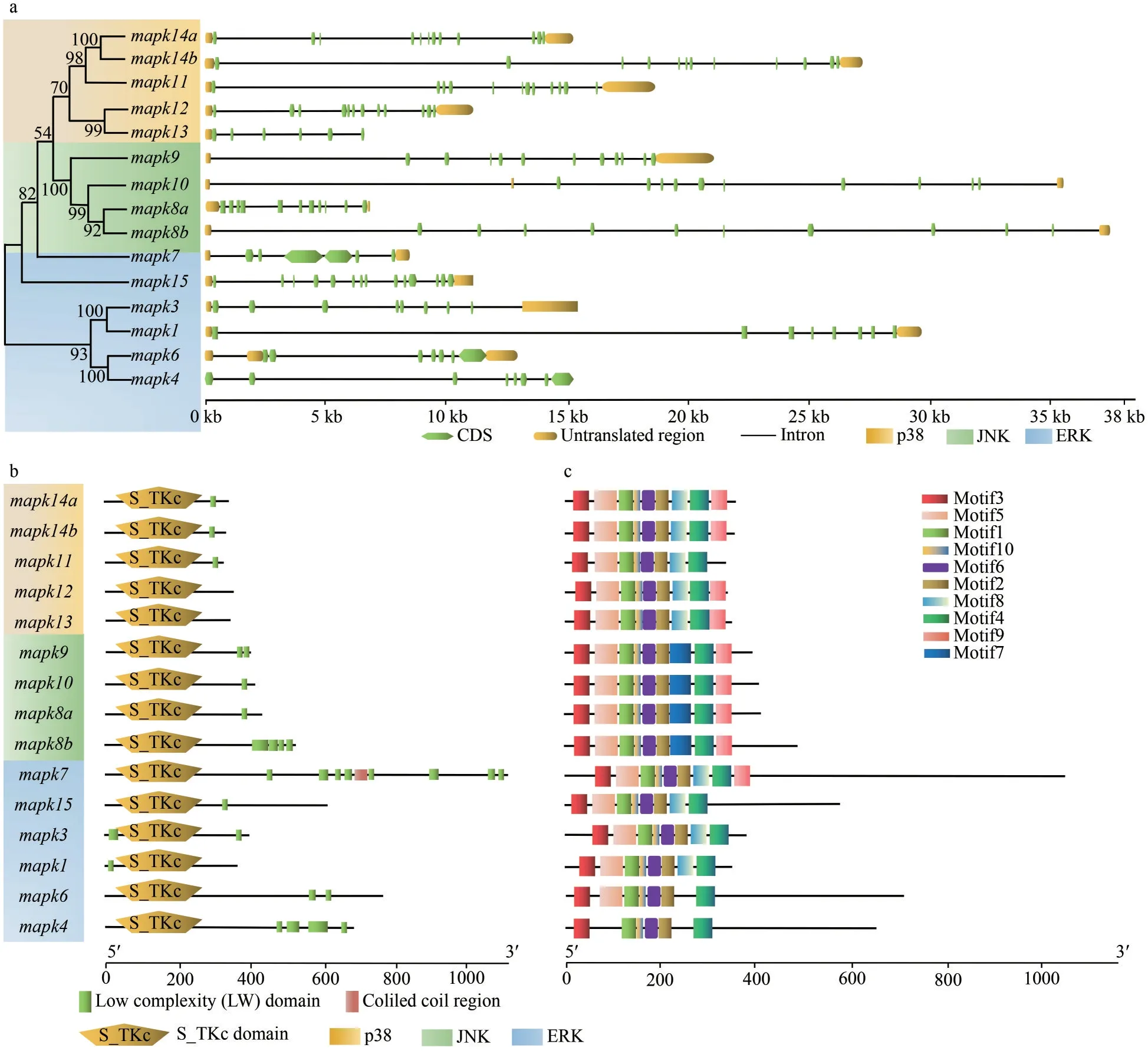
Fig.5 Gene structure, functional domain, and conserved motif analyses of mapk genes in black rockfish
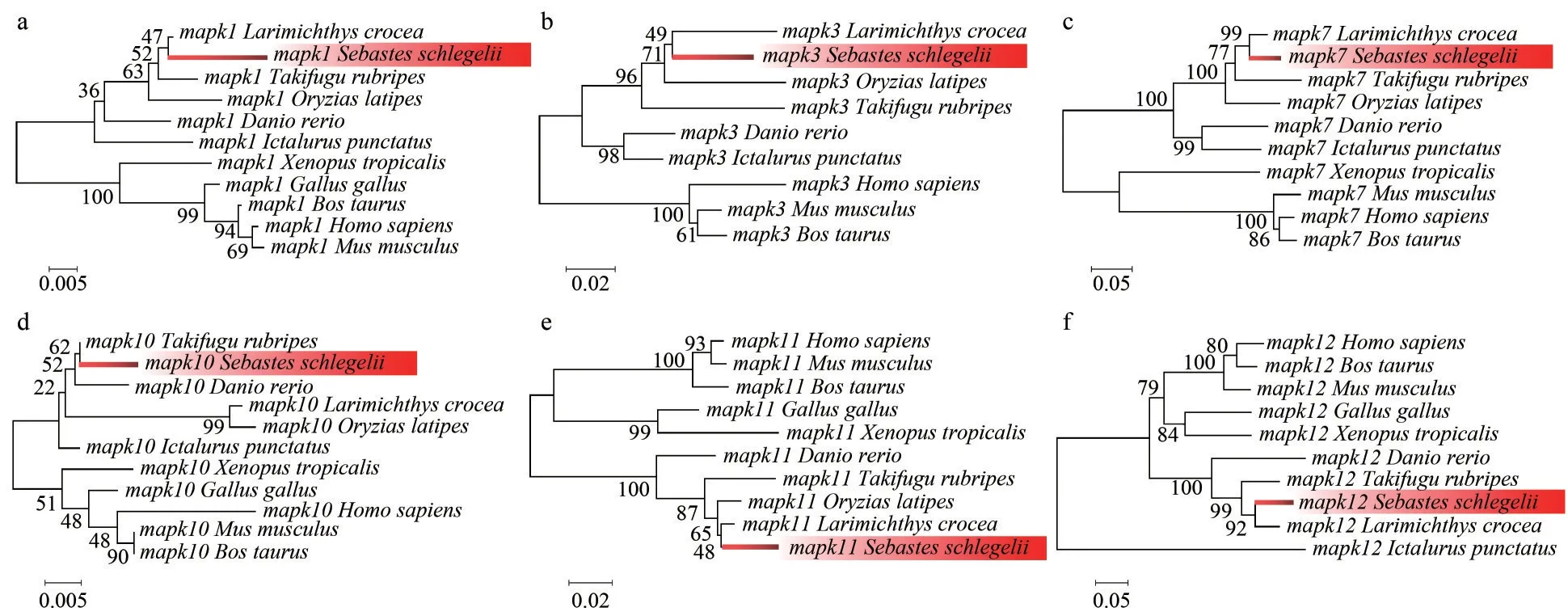
Fig.6 Phylogenetic tree of mapk genes for branch-sites model
Totally, 15mapkgenes were systematically identified and functionally characterized in black rockfish, which were further divided into three subfamilies: ERKs, JNKs, and p38s.Similar to previous results of other vertebrates, members of ERK, JNK, and p38 subfamilies in black rockfish possess different dual-phosphorylation sites,including TEY, TPY, and TGY, all are located in the regions of activation loop.The three major clades of the phylogenetic tree are consistent with the classification of subfamilies, suggesting their high conservation in evolution.Moreover, the evolutionary conservation ofmapkgene family was further confirmed by the comprehensive analysis of syntenic region, gene structure, protein domain, and chromosomal distribution.The copy numbers ofmapkgenes in the selected species are relatively conversed and most of them range from 13 to 15.It has been reported that an extra third round of the whole-genome duplications (WGD) occurred in teleost fish, which may lead to duplication or lose some duplicates during this process (Watterson,1983; Meyer and van de Peer, 2005).Interestingly,the duplicatedmapk8andmapk14genes were found in teleost only.We thus confirmed that they are teleost-specific, and speculated that whole-genome duplication or recombination events contributed to their duplication, which is supported by previous studies (Vogel et al., 2005; Glasauer and Neuhauss,2014).To better understand the changes to which thesemapkgenes have been subjected during evolution, selective pressure analysis was conducted.Totally, 10 positive selection sites were identified inmapk1(1 site),mapk3(2 sites),mapk7(1 site),mapk10(1 site),mapk11(3 sites), andmapk12(2 sites).These genes and sites under positive selection pressure, which may be related to the adaptive evolution of black rockfish.However,further exploration is still needed to clarify the functions of the positively selected amino acid sites in thesemapkgenes.However, the positively selected amino acid sites in these genes affect or alter their specific biological functions remains to be further explored.
Due to the key roles ofmapkgenes in almost all aspects of growth, development and response to environment stimuli, the expressions ofmapkgenes in different development stages and tissues have been well characterized in many species, such as zebrafish (Krens et al., 2006), seahorse(Hippocampus erectus) (Wang et al., 2021), Yesso scallop (Patinopecten yessoensis) (Sun et al., 2016),and Pacific white shrimp (Litopenaeus vannamei)(Yan et al., 2013).In black rockfish,mapkgenes displayed different expression patterns in the 8 tested tissues, implying their diverse physiological functions among tissues.Almost allmapkgenes were expressed at relatively high levels in brain.The result demonstrated that themapkgenes may be closely associated with the complex patterns of signal transduction in the brain of black rockfish(Kaminska et al., 2009; Zhou et al., 2015).High expressions ofmapk1,mapk13, andmapk14agenes were detected in main osmoregulatory tissue of gill,suggesting they may participate in the response to osmotic stresses.Especially,mapk14ais important for the adaptation to hyper-osmotic stress in the gill of spotted sea bass (Tian et al., 2019).In addition,gill along with skin and intestine are the main mucosal tissues of fish, which are the primary sites for host-bacteria interaction and key components of the innate immune system (Gomez et al., 2013; Liu et al., 2016).The abundant expression of thesemapkgenes in these mucosal tissues reflected the defense mechanisms of innate immunity against bacteria or other pathogens in black rockfish.
Edwardsiella piscicidais a common and important pathogen causing gastrointestinal and systemic infections to aquatic animals, especially fish (Schlenker and Surawicz, 2009; Leung et al.,2012).Similarly,E.piscicidaalso causes the disease of black rockfish with economic losses.Themapkgene has been shown the ability as regulator of innate and adaptive immune responses.Therefore,the potential roles ofmapkgenes in immune response of black rockfish were investigated via detecting the expression levels ofmapkgenes afterE.piscicidainfection.In this study, the expression levels of severalmapkgenes in gill, skin, and intestine tissues were significantly changed afterE.piscicidainfection, suggesting their important roles in mucosal immunity.In detail, the p38mapkgenes, includingmapk12(p38γ),mapk13(p38δ),mapk14a(p38αa), andmapk14b(p38αb),were all up-regulated at all the time points in the examined tissues, except formapk14ain skin.Similarly, the expression of p38mapkgenes was also induced by bacterial or pathogen simulation in mucosal tissues of grouper (Epinephelus coioides) (Sun et al., 2017),Chinese shrimp (Fenneropenaeus chinensis) (He et al., 2018), and grass carp (Ctenopharyngodon idella) (Sun et al., 2020).Evidence has been demonstrated that p38mapkcan help host in preventing bacterial infection in aquatic invertebrates and vertebrates (Hansen and Jørgensen, 2007; Uribe et al., 2011; Sun et al., 2020).Moreover, p38mapkgenes can be phosphorylated by transcription factor and translational elements, then inflammatory responses be activated, and can regulate the expressions of numerous pro-inflammatory cytokines such asTNF-αand interleukins (Ils) (Lee et al.,1994; Raingeaud et al., 1995; Zarubin and Han,2005; Hansen and Jørgensen, 2007; Sun et al., 2019;Zhang et al., 2019).Moreover, we found thatE.piscicidainfection could significantly induce the expression ofmapk1(erk2) andmapk15(erk7/8) in mucosal tissues.The results confirmed the function of ERKmapkgenes in bacterial-mediated immune response in black rockfish.So far, ERK studies of immune response were mainly focused on mainlyerk1anderk2genes.The up-regulated expression oferk2gene was observed in the gill of Chinese shrimp (Li et al., 2013) and grouper (Sun et al.,2018).More importantly, inhibition toerk1/erk2signaling in zebrafish would diminish the host antibacterial defense and stimulate the pathogenesis ofE.piscicida, providing direct evidence thaterk1/erk2signaling can play vital roles in response toE.piscicidainfection (Yang et al., 2007).However,the understanding oferk7/8gene and their biological function in mucosal tissues are still limited.
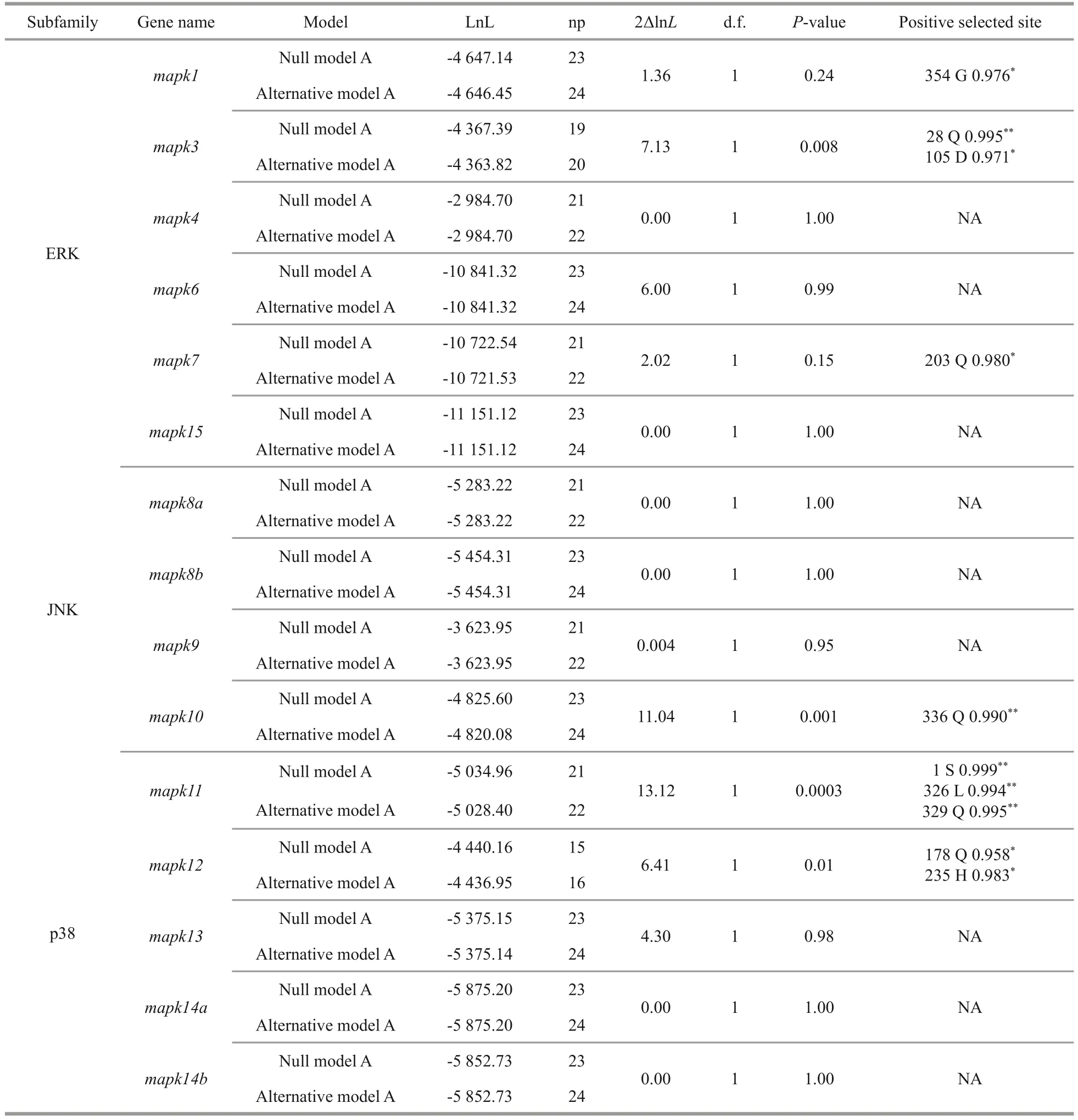
Table 2 The parameters and statistical significances of likelihood ratio tests using the branch-site model
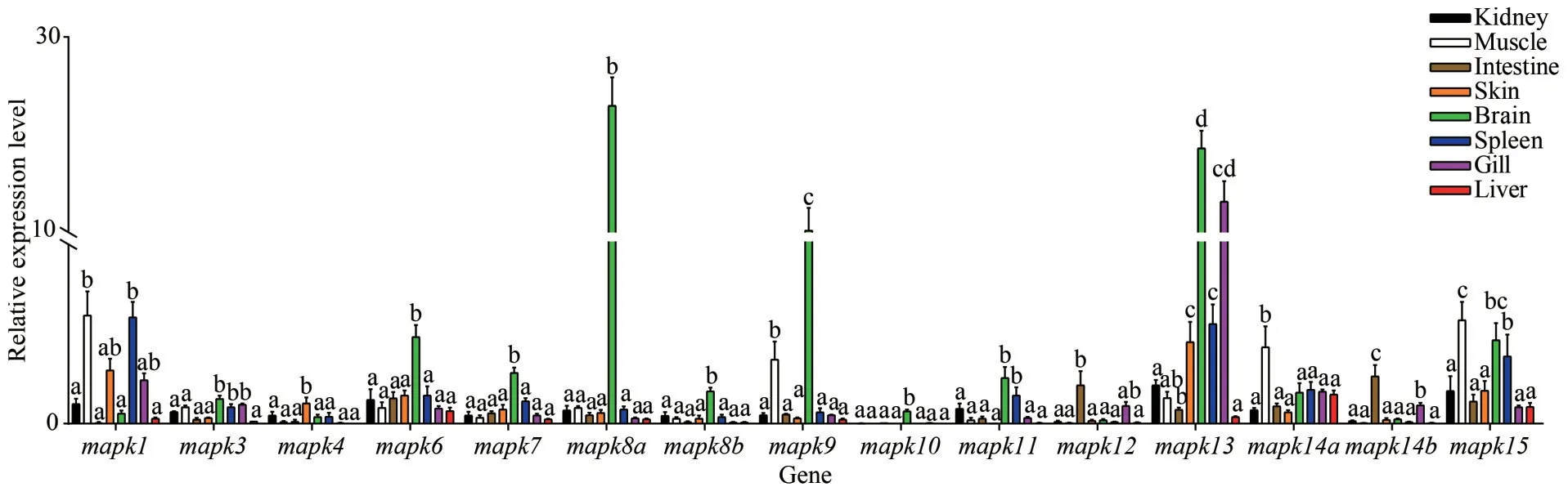
Fig.7 Expression patterns of mapk genes in healthy tissues of black rockfish

Fig.8 Expression patterns of mapk genes in intestine, skin,and gill at different time points after A. salmonicida infection
5 CONCLUSION
Totally, 15mapkgenes were systematically identified in black rockfish, and phylogenetic positions, syntenic analysis were performed to provide additional evidence for their annotations.Ten sites in 6mapkgenes (mapk1,mapk3,mapk7,mapk10,mapk11, andmapk12) were found to undergo significantly positive selection in black rockfish.Moreover, the results of qRT-PCR show thatmapkgenes displayed distinct expression patterns in different tissues.The expression patterns ofmapk12,mapk13,mapk14a,mapk14b, andmapk15were significantly changed in mucosal tissues afterE.piscicidainfection, indicating the important roles ofmapkgenes against bacterial infection in black rockfish.However, further studies are still needed to characterize the immune response mechanism ofmapkgenes in teleost.
6 DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Trends of carbon and nutrient accumulation through time in the Andong salt marsh, Hangzhou Bay, China*
- Physical processes determining the distribution patterns of Nemopilema nomurai in the East China Sea*
- Comparison in structure and predicted function of epiphytic bacteria on Neopyropia yezoensis and Neopyropia katadae*
- Interaction between macroalgae and microplastics: Caulerpa lentillifera and Gracilaria tenuistipitata as microplastic bio-elimination vectors*
- Lake regime shift from submerged macrophyte to phytoplankton affected phosphorus speciation in sediment and eutrophic state in Caohai Lake, Guizhou, China*
- Temporal characteristics of algae-denitrifying bacteria co-occurrence patterns and denitrifier assembly in epiphytic biofilms on submerged macrophytes in Caohai Lake, SW China*
