Study on the mechanism of Fuzi in the treatment of allergic rhinitis based on network pharmacology and experimental validation
2023-12-06LILinDINGShunXUZhengyangYANJingrenZHANGQimengMUZhonglin
LI Lin, DING Shun, XU Zheng-yang, YAN Jing-ren, ZHANG Qi-meng, MU Zhong-lin
Department of Otolaryngology, Head and Neck Surgery, the First Affiliated Hospital of Hainan Medical University, Haikou 570102, China
Keywords:
ABSTRACT Objective: To study the key target genes and signaling pathways in the treatment of Allergic Rhinitis(AR) with Radix Aconiti Lateralis Preparata (aka Fuzi).Methods: The TCMPS and PubChem databases were used to screen the active ingredients and target genes of Fuzi using oral bioavailability and drug similarity as screening conditions, and the GeneCards database was used to screen the target genes of AR.The online tool Venny2.1 was used to screen the target genes of Fuzi for the treatment of Allergic Rhinitis; the STRING database was used to obtain the protein-protein interaction (PPI) network of drug-disease targets, and the key target genes were identified by the MCC algorithm.The potential biological processes and signaling pathways were identified by GO enrichment and KEGG enrichment analysis.Finally,animal experiments were conducted to demonstrate the therapeutic effect ofFuzi on Allergic Rhinitis.Results: The TCMSP, PubChem and GeneCards databases were used to screen the 21 active compound components of Fuzi and 68 potential therapeutic target genes of Fuzi for Allergic Rhinitis.PPI network analysis identified the top ten key target genes, namely:PTGS2, TNF, IL6, AKT1, ALB, STAT3, CCL2, CXCL8, VEGFA and JUN, GO functional and KEGG pathway enrichment analysis showed that the significantly enriched functions and pathways of Fuzi on Allergic Rhinitis were closely related to Allergic Rhinitis.Finally, animal experiments were conducted to verify that Fuzi is effective in the treatment of Allergic rhinitis.Conclusion: Increased expression of IL-6 and TNF-α in nasal mucosal tissues of patients with Allergic Rhinitis was positively correlated with indicators related to the disease activity of AllergicRhinitis.Fuzi ameliorated the inflammatory changes in mice with Allergic Rhinitis by inhibiting the activation of Toll-like signaling pathway in the nasal mucosa and decreasing the expression activity of IL-6 and TNF-α.
1.Introduction
Allergic rhinitis (AR) is one of the most common diseases in otolaryngology and is a symptomatic, inflammatory and immune disease of the nasal mucosa[1].Allergic rhinitis can reduce the quality of life of patients and lead to the development of other diseases such as asthma and obstructive sleep apnea[2], and about 80% of patients with allergic rhinitis have clinical symptoms before the age of 20 years and reach a peak between 20 and 40 years of age, followed by a gradual decline[3].Allergic rhinitis is a multifactorial disease with a complex pathogenesis that involves multiple inflammatory cells and inflammatory mediators[4].Current treatment options for allergic rhinitis include patient education, allergen avoidance,pharmacotherapy, immunotherapy, Chinese acupuncture, Chinese herbal formulas, and surgery[5].Western pharmacological treatment is currently the predominant treatment modality for allergic rhinitis,but these drugs are often prone to side effects such as fatigue,hormone resistance, and sedation[6].Therefore, there is a need for an effective and safe drug to prevent and treat allergic rhinitis[7].Since ancient times, Fuzi has been a representative herbal medicine for clinical treatment of allergic rhinitis, and Fuzi has been used for calming asthma, improving immune function, antioxidant,dispersing cold and relieving pain, and anti-cold and antibacterial effects[8, 9].However, the specific mechanism of action of Fuzi in the treatment of allergic rhinitis is still to be determined.Traditional mechanism studies based on a single component cannot achieve a comprehensive and systematic understanding of the pharmacological effects of Chinese medicine, and network pharmacology with its holistic and systematic characteristics has been widely used in the study of Chinese medicine, and it is feasible to apply network pharmacology to study the mechanism of Fuzi in the treatment of allergic rhinitis.
In this study, we applied a network pharmacology approach to obtain the main active compound components and target genes of Fuzi for the treatment of allergic rhinitis, and screened the potential biological processes and signaling pathways of Fuzi for the treatment of allergic rhinitis by enrichment analysis.On this basis, a mouse model of allergic rhinitis was established to observe the therapeutic effects of Fuzi on allergic rhinitis.This will lay the foundation for further research on the pharmacological mechanism of Fuzi in the treatment of allergic rhinitis.
2.Materials and Methods
2.1 Database and software analysis
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) database was used to screen for bioactive components with OB 20% and DL 0.1 as the screening criteria.The active compound components related to Fuzi were found[10].Pharmacokinetics (Absorption, Distribution,Metabolism, and Eexcretion, ADME) is an important limiting factor for screening the bioactive compound components of Fuzi, which refers to the absorption, distribution, metabolism, and excretion of drug compounds.In ADME, Oral Bioavailability (OB) and Druglikeness (DL) are the most important pharmacokinetic properties.OB is the rate and extent of oral drug absorption from the gastrointestinal tract into the circulatory system [11].DL is a qualitative concept of drug setting and can be used as a herbal DL is a qualitative concept of drug setting and can be used as a basis for the selection of “druglike” components.
2.2 Genetic screening of the active targets of Fuzi
The target genes corresponding to the main active compounds of Fuzi were obtained by screening the active compounds of Fuzi in the TCMSP database, setting the screening species category through the PubChem database[12], and transferring the target protein names to“gene symbol”.Then, the UniProt database was used to standardize the gene names[13], and the target genes corresponding to the active compounds of Fuzi were obtained after removing duplicate genes.
2.3 Target gene screening for allergic rhinitis
The GeneCards database is a database of human genetic information[14], and target genes associated with allergic rhinitis were searched in the GeneCards database by using the term “allergic rhinitis”.
2.4 Acquisition of overlapping target genes for allergic rhinitis and Fuzi
The overlapping genes of Fuzi and allergic rhinitis were screened by the online tool Venny 2.1[15], a commonly used statistical chart which is a graphical representation of the overlapping regions of a collection of elements.The target genes of Fuzi for the treatment of allergic rhinitis are these screened overlapping genes are shown by using the Wayne diagram.
2.5 Construction of “Compound-Gene-Disease” network diagram
The database was used to screen the target genes of Fuzi for the treatment of allergic rhinitis, and a Compound-Gene-Disease(C-G-D) network graph was constructed using Cytoscape software for visualization, and a network analysis tool was used to derive the connectivity of each node, which indicates the number of nodes directly connected to the node.The greater the connectivity, the more important the role of the node in the network[16].
2.6 Protein interaction network graph construction
Protein-protein interaction (PPI) network was studied by STRING database for the overlapping genes of Fuzi and allergic rhinitis, and the PPI network graph was constructed at a confidence level of 0.4[17].The PPI network generated based on the database information was imported into Cytoscape software, and then the top 10 most core genes, which are the top 10 important pivotal genes of Fuzi for the treatment of allergic rhinitis, were screened based on the MCC algorithm using the CytoHubba plug-in in Cytoscape[18].
2.7 Enrichment analysis
GO functional databases can help us to better understand gene functions such as Cellular Components (CC), Biological Processes(BP) and Molecular Functions (MF)[19].KEGG functional databases are databases from which gene and genomic information can be The DAVID database is an online analysis software.In order to explore the specific roles of target proteins in terms of gene functions and signaling pathways, GO functional enrichment analysis and KEGG signaling pathway enrichment analysis were performed using the functional annotation tool in the DAVID database for the active compound components of Fuzi and allergic rhinitis interaction target genes[21, 22], the top ten keywords of GO functional enrichment analysis regarding cellular components, biological processes and molecular functions were selected and the results were visualized by bar graphs.The top 20 key pathways of KEGG were selected and setP<0.05, and the results were visualized by analyzing the plots.
2.8 Experimental validation
2.8.1 Experimental materials and methods
(1) Experimental drugs
100 g of Fuzi was purchased from Haikou Traditional Chinese Medicine Hospital, soaked in 10 times the amount of deionized water for 10 min, boiled on military fire and decocted on civil fire for 2 h, filtered to obtain the first decoction; the second decoction was added to 8 times the amount of deionized water, boiled on military fire and decocted on civil fire for 1.5 h, filtered to obtain the second decoction, and the two decoctions were combined and concentrated to a raw drug amount of 1.52 g·mL-1concentrate, which was placed in a refrigerator at -80 ℃ for storage.IL-6 and TNF-α antibodies were purchased from Absin (Shanghai) Biotechnology Co.
2.8.2 Animal model construction
(1) Female mice aged 4-6 weeks, weighing approximately (20±2) g,were purchased from Fengdong New City Experimental Zoo, Xi’an New District, and were housed in an SPF-grade animal laboratory:temperature (22±2) ℃, humidity (60±5) %, 12 h light/dark cycle,and all experiments were performed in accordance with the Chinese National Guide for the Care and Use of Laboratory Animals.The study was approved by the Ethics Committee of Hainan Medical College (animal ethics number: HYLL-2021-382).
(2) After 1 week of acclimatization feeding, 30 female mice were randomly and equally divided into Normal Control group (NC group), Allergic Rhinitis group (AR group) and Radix Aconiti Lateralis group (Fuzi group).Preparata group, Fuzi group).According to the references, with appropriate adjustments we created the following mouse model of allergic rhinitis[23].During the first 14 d, mice in the Fuzi group and the allergic rhinitis group were sensitized by intraperitoneal injection of a sensitizing solution consisting of ovalbumin (OVA) 40 µg and Al(OH)31.5 mg dissolved in saline to make a total of 0.2 mL of sensitizing solution, which was administered once every 2 days from day 15 to day 21 at the rate of 40 µL of 5% OVA nasal drops per mice were stimulated bilaterally in the nasal cavity once a day from day 15 to day 21.From day 22 to day 30, mice in the Fuzi group were gavaged at a dose of 1.56 g/kg once daily, while the allergic mice and Fuzi group were stimulated with OVA nasal drops every other day to maintain nasal stimulation,and mice in the normal group were replaced with the same volume of saline.
2.8.3 HE staining
The intact nasal mucosa of each group of mice was soaked in 4%paraformaldehyde water for 24 h.After dehydration, wax immersion,embedding and sectioning, the paraffin-embedded nasal mucosa tissue was stained with hematoxylin and eosin method, and light microscopy was used to evaluate and record the histopathological changes.
2.8.4 Protein immunoblotting assay
Proteins were isolated from each group of nasal mucosal tissues for identification of IL-6 and TNF-α expression.The extracted proteins were added to BSA standard assay solution for protein concentration identification and the proteins were separated by SDS-PAGE gel electrophoresis.The proteins were then transferred to transfer membrane filter paper and then incubated with 5% skim milk for 2 hours before adding primary antibody and incubating overnight at 4 ℃ in the refrigerator.Afterwards, the membrane is washed with TBST and the secondary antibody solution is added and incubated for 2 h at room temperature.Finally, the membrane is developed by a developing instrument and the target band/corresponding relative expression level is measured.
2.8.5 Immunohistochemical assay
Dewaxed paraffin sections are dehydrated by gradient in xylene and alcohol.Antigens were healed by immersion in buffer at 90°C for 6 min, followed by incubation with 3% H2O2for 10 min to inactivate endogenous peroxidase.Then BSA was added dropwise to block histones, primary antibodies to TNF-α and IL-6 were added at 4℃, followed by incubation with secondary antibodies dropwise for 20 min, then horseradish enzyme-labeled streptavidin working solution was added for 15 min, and color development solution and hematoxylin solution were added.After blocking was performed, an inverted microscope was used for observation.
2.8.6 Statistical analysis
Descriptive statistical analysis was used to statistically describe the experimental results.t-test analysis was used between two groups and one-way ANOVA was used between multiple groups,and P<0.05 was considered statistically significant.All statistical analyses were performed using GraphPad Prism 8.
3.Results
3.1 Screening of target genes for the action of Fuzi
In the TCMSP database, OB and DL were used as the screening conditions for target genes to find 21 active compound components of Fuzi.The details of the 21 biologically active compound components of Fuzi are shown in Table 1.203 action target genes of Fuzi were then obtained by the PubChem database after eliminating 64 duplicate genes.
3.2 Screening of allergic rhinitis target genes and overlapping target genes
The GeneCards database was used to screen 1832 target genes related to allergic rhinitis after removing duplicate genes, and then Venny 2.1 software was used to establish the connection between 203target genes of Fuzi and 1832 target genes of allergic rhinitis, and a total of 68 overlapping genes were screened, and the Venn diagram of overlapping target genes of allergic rhinitis and Fuzi is shown in Figure 1, which is the target genes for the treatment of allergic rhinitis.

Tab 1 Information on 21 Fuzi compounds in detail.OB: oral bioavailability.DL:drug-likeness.
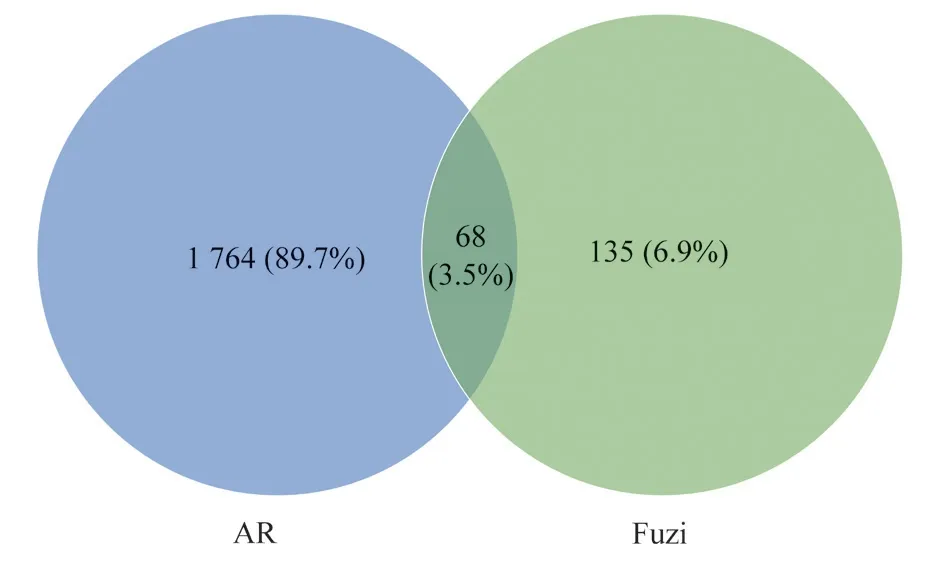
Fig 1 Allergic Rhinitis and Fuzi targets depicted in a Venny diagram.
3.3 Construction of “Compound-Gene-Disease” Network Diagram
Compound-gene-disease” network diagram is a valuable tool to study the basic process of Chinese medicine treatment.The“compound-gene-disease” network diagrams of allergic rhinitis and Cyperus rotundus were constructed using Cytoscape software as shown in Fig.2, with the red polygonal nodes in the middle indicating diseases and the light green rectangular nodes indicating the main active compounds of Cyperus rotundus including deoxyandrographolide ( The dark green rectangular nodes indicate the overlapping genes of P.Fuzi and allergic rhinitis, such as PTGS2,TNF, ALB, CXCL8, IL6, VEGFA, JUN, etc, JUN, etc.Each active compound is connected to multiple targets, and multiple targets are also connected to multiple components.This network diagram can show that Fuzi can act on multiple targets through multiple compound components in it, thus acting as a treatment for allergic rhinitis.

Fig 2 “Compound-Gene-Disease” Network Diagram
3.4 Construction of PPI network graph
A PPI network graph of overlapping genes of Fuzi and allergic rhinitis was constructed using STRING database, see Figure 3,which includes 68 nodes and 860 connections, and then the top 10 overlapping pivotal genes of Fuzi for the treatment of allergic rhinitis were screened using the MCC algorithm method in CytoHubba plugin mainly as PTGS2, TNF, IL6, AKT1, AL8, STAT3, CCL2,CXCL8, VEGFA, JUN, and the network diagram of the top 10 overlapping hub genes was constructed as shown in Figure 4, which are the key genes of Fuzi for the treatment of allergic rhinitis.
3.5 GO functional enrichment analysis
The 68 overlapping target genes were entered into the DAVID database for GO function enrichment analysis, which can illustrate the biological functions of various potential targets of Fuzi for the treatment of allergic rhinitis.The GO function enrichment analysis is shown in Figure 5, in which the GO function enrichment in BP, the top 10 relevant entries were selected, mainly involving transcriptional positive regulation, nitric oxide biological The top 10 entries were selected for positive regulation of transcription, positive regulation of nitric oxide biosynthesis, positive regulation of smooth muscle cell proliferation, positive regulation of tumor necrosis factor production, etc.In CC, the top 10 relevant entries were selected,mainly for cell surface, plasma membrane, protein complex and apical plasma membrane involved in major cellular components.In MF, the top 10 relevant entries were selected, mainly including DNA binding to transcriptional regulatory regions, binding of transcription factors, sequence-specific DNA binding to the proximal region of core promoters, and binding of protein extracellular matrix.
3.6 KEGG functional enrichment analysis
In order to explore the pathways of action of the active compound components of Fuzi through target genes for the treatment of allergic rhinitis, the common targets of Fuzi for the treatment of allergic rhinitis were mainly enriched in 20 pathways (P<0.05), and the KEGG functional enrichment analysis was further constructed,as shown in Figure 6, including TNF signaling pathway, T cell receptor signaling pathway, Toll-like receptor signaling pathway,HIF-1 signaling pathways.The Toll-like receptor signaling pathway map was selected for display, see Figure 7, where green represents common genes, combined with the core target genes in the PPI network map containing IL-6 and TNF-α hub genes in the Tolllike receptor signaling pathway, so we selected this pathway for subsequent study.
3.7 Effect of Fuzi on histopathological changes in the nasal mucosa of mice with allergic rhinitis
The results of the histological changes of the nasal mucosa of mice in each group by HE staining are shown in Figure 8.The normal mice group showed neatly arranged cells with intact cell structure.Compared with the normal group, the nasal mucosa of the allergic rhinitis group showed different degrees of disruption and collapse of mucosal integrity, fibrous tissue proliferation and inflammatory cell infiltration.In contrast to the allergic rhinitis group, the above pathological changes were significantly reduced in the mice in the Fuzi group.

Fig 4 The top 10 hub gene network of Fuzi therapy in response to Allergic Rhinitis were analyzed using MCC

Fig 5 Evaluation of targets based on the GO
Tab 2 Statistical table of grayscale values of IL-6 protein expression in nasal mucosal tissue of mice in each group (n=10, ±s)

Tab 2 Statistical table of grayscale values of IL-6 protein expression in nasal mucosal tissue of mice in each group (n=10, ±s)
组别 images/BZ_7_278_350_295_382.png±s d正常对照组 0.7502±0.0596变应性鼻炎组 1.3355±0.0914附子组 0.8415±0.0247 F 71.312 P<0.001
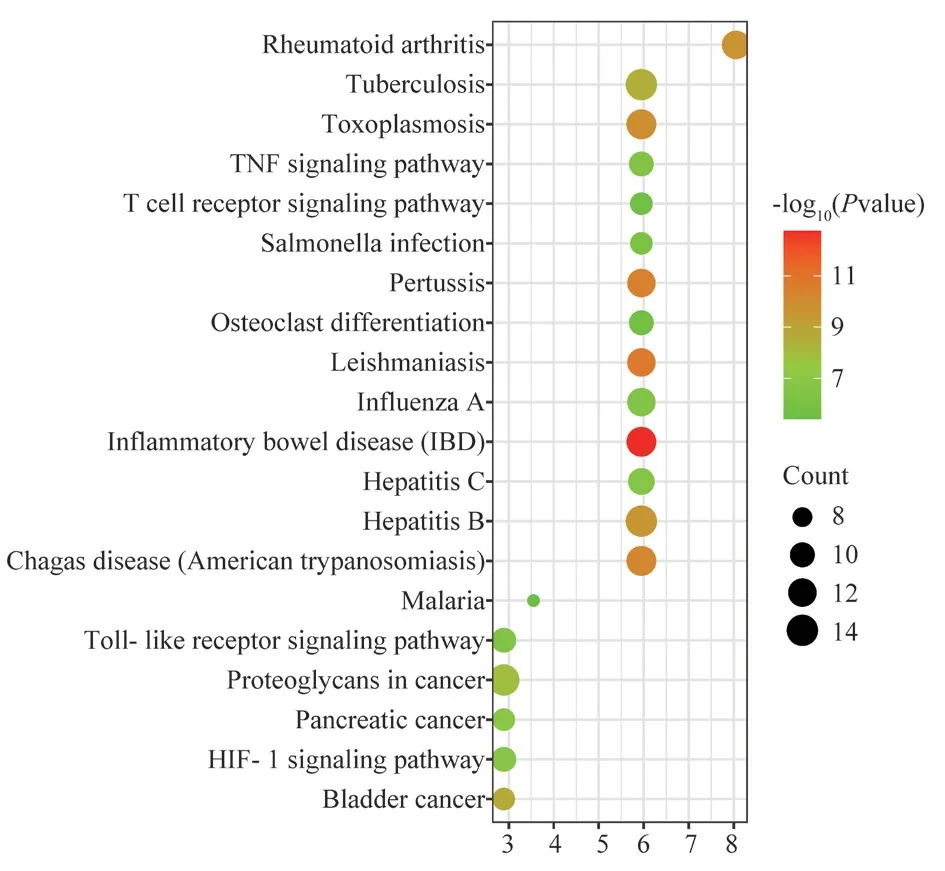
Fig 6 KEGG functional enrichment analysis diagram
3.8 Protein expression levels of IL-6 and TNF-a detected by protein immunoblotting
The WB results of the expression levels of IL-6 and TNF-α in the nasal mucosa of mice in each group are shown in Figure 9.The differences were statistically significant (P<0.000 1; P<0.000 1).The expression of IL-6 and TNF-α protein in the nasal mucosa of mice in the allergic rhinitis group was higher than that in the Fuzi group, and the differences were statistically significant (P=0.000 2;P<0.000 1).The expression of IL-6 in the nasal mucosa of mice in the Fuzi group was not statistically different from that of the normal control group (P=0.269 7); the expression of TNF-α in the nasal mucosa of mice in the Fuzi group was statistically different from that of the normal control group (P=0.000 1).The above results indicated that IL-6 and TNF-α were present and expressed in the nasal mucosa tissues of mice, and that Fuzi could reduce the inflammatory response by regulating the expression of IL-6 and TNF-α, which in turn confirmed the results of the network pharmacological analysis.

Fig 7 Toll-like receptor signaling pathway
Tab 3 Statistical table of grayscale values of TNF-α protein expression in nasal mucosal tissue of mice in each group (n=10,±s)

Tab 3 Statistical table of grayscale values of TNF-α protein expression in nasal mucosal tissue of mice in each group (n=10,±s)
组别 images/BZ_7_278_350_295_382.png±s d正常对照组 0.2719±0.0251变应性鼻炎组 0.8658±0.0292附子组 0.5311±0.0369 F 280.532 P<0.001
Tab 4 Statistical table of the expression level of IL-6 in the nasal mucosa tissue of mice in each group (n=10, ±s)

Tab 4 Statistical table of the expression level of IL-6 in the nasal mucosa tissue of mice in each group (n=10, ±s)
组别 images/BZ_7_278_350_295_382.png±sd正常对照组 0.20±0.05变应性鼻炎组 0.53±0.12附子组 0.28±0.17 F 41.683 P<0.001
3.9 Detection of protein expression levels of IL-6 and TNF-a by immunohistochemistry
In order to confirm the validity of the network pharmacological data, immunohistochemical assays were performed on the nasal mucosa of each group of mice, and the statistical tables of the expression levels of IL-6 in the nasal mucosa tissues of each group of mice are shown in Tables 4 and 5, respectively.the results of immunohistochemical assays for the expression levels of IL-6 and TNF-α in the nasal mucosa of each group of mice are shown in Figure 10.most of the IL-6 and TNF-α protein-positive particles were present in the cell membrane and cell pulp, and They were brown or brownish yellow in color.The nasal mucosal Fuzi was undamaged and no inflammatory cell infiltration was seen in the normal group, while the glandular and mucosal epithelial staining was darker and the nasal mucosal Fuzi was damaged in the allergic rhinitis group, and the damage to the nasal mucosa was significantly lighter, with less glandular hyperplasia and less inflammatory cell infiltration in the Fuzi group.The results showed that the positive expression of IL-6 and TNF-α in the nasal mucosa of mice in the allergic rhinitis group was higher than that in the normal control group, which was statistically significant (P<0.001; P<0.001).The positive expression of IL-6 and TNF-α in the nasal mucosa of mice in the group was higher than that in the normal control group, which was statistically significant (P<0.05;P<0.05).The expression of IL-6 and TNF-α in the group was lower than that in the allergic rhinitis group, which was statistically significant (P<0.001;P<0.05).The results indicated that Fuzi improved the inflammatory response of nasal mucosa in mice.

Figu 9 WB results of IL-6 and TNF-α expression levels in the nasal mucosa of mice in each group.(A) Detection of IL-6 and TNF-α expression in the nasal mucosa of mice in each group by WB method.(B,C) Statistical analysis of IL-6 and TNF-α protein expression levels in the nasal mucosa of mice in each group (*P<0.05, **P<0.01, ***P<0.001).

Fig 10 Immunohistochemical detection of IL-6 and TNF-α expression levels in the nasal mucosa of mice in various groups.(A)Expression of IL-6 and TNF-α in the nasal mucosa of mice in each group.(B,C)Statistical analysis of IL-6 and TNF-α protein expression levels in the nasal mucosa of mice in each group (*P<0.05, **P<0.01, ***P<0.001).
Tab 5 Statistical table of the expression level of TNF-α in the nasal mucosa tissue of mice in each group (n=10, ±s)

Tab 5 Statistical table of the expression level of TNF-α in the nasal mucosa tissue of mice in each group (n=10, ±s)
组别 images/BZ_7_278_350_295_382.png±sd正常对照组 0.24±0.08变应性鼻炎组 0.59±0.08附子组 0.32±0.10 F 43.732 P<0.001
4.Discussion
Allergic rhinitis is primarily an inflammatory rhinopathy mediated by IgE following exposure to allergens in atopic individuals[24].In atopic individuals, exposure to indoor and outdoor allergens(including pollen, mold, dust mites, and animal dander) may promote the production of antigen-specific IgE.Reintroduction of allergens triggers early and late reactions leading to the clinical manifestations of allergic rhinitis[25].The multi-component, multitarget, and multi-pathway properties of TCM and the high cure rate and low cost of TCM treatment[26] have led to an increasing number of people choosing to use TCM therapies for the treatment of allergic rhinitis[27].From a TCM perspective, TCM believes that the nose can reflect the physiological and pathological manifestations of the lungs.As external evils (considered pathogenic factors in TCM) invade the body through the mouth and nose, they invade the nasal cavity and impair the declination function of the lung,leading to the imbalance of fluids and causing body fluids to become phlegm and mucus.This eventually leads to the formation of allergic rhinitis [28].Allergic rhinitis mostly originates from deficiency of positive qi, and in treatment, the main principle has been to benefit qi and warm yang, and Fuzi has been a typical herbal medicine for clinical treatment of allergic rhinitis in weak cold syndrome since ancient times[29,30]; Fuzi is pungent, hot, and warming to the lining and helps shao yin and yang qi[31], and has functions such as improving immunity and anti-inflammatory and antioxidant[32].On the other hand, the mechanism of action of Fuzi in the treatment of allergic rhinitis is still unclear, and the application of network pharmacology can investigate the complexity of the components, targets, pathways, and modes of action of herbs or herbal prescriptions[33] to more accurately predict the target genes and the main This study used network pharmacology to elucidate the potential physiological processes of Fuzi in the treatment of allergic rhinitis.A total of 21 bioactive compound components and 203 target genes were screened under the screening conditions of OB 20% and DL 0.1.The bioactive compound components included Deoxynivalenolide, Karanjin and hypaconitine, which are the main active compounds of Fuzi for the treatment of allergic rhinitis.Deoxyandrographolide contains diterpene lactones, flavonoids, and polyphenols[34], which have been shown to have anti-inflammatory properties in many studies[35], and deoxyandrographolide has the basic structure of diterpene lactones and a five-membered lactone ring exerting anti-inflammatory effects[36].Karanjin is an active furanoflavone and flavonoids are anti-inflammatory factors.Studies in rat models have shown that hydroxanthin has anti-inflammatory properties by blocking H+and K+ATPases[37].Hypaconitine protects epithelial cells from oxidative stress[38].In mice, hypaconitine has been shown to have potent anti-inflammatory effects, inhibiting cyclooxygenase-2, tumor necrosis factor, interleukin-1, and prostaglandin E2 production[39].
Based on the importance of each target gene, we selected the top 10 important pivotal genes PTGS2, TNF, IL6, AKT1, ALB, STAT3,CCL2, CXCL8, VEGFA, and JUN.PTGS2 is a key enzyme in the production of prostaglandin family, which is the target of action of NSAIDs and is now known to be involved in cellular inflammation,apoptosis, and other physiological activities, PTGS2 transcription is triggered by several cytokines associated with inflammatory signaling pathways, including nuclear factor B, activator protein 1,and CCAAT enhancer binding protein[40].TNF-α is a member of the TNF family that regulates cell death and promotes cytoplasmic membrane rupture leading to the release of inflammatory factors[41].Il-6 limits neutrophil production and increases granulocyte death.In allergic rhinitis, changes from rapid host defense to adaptive sustained systemic inflammation are associated with the pro-inflammatory effects of IL-6.AKT1 is an important component of the P13K signaling pathway and promotes nasal mucosal cell survival and proliferation[42].CCL2 stimulates migration and adhesion of monocytes and macrophages at the site of injury and triggers a series of inflammatory responses when CCL2 comes in contact with its receptors.CXCL8 is involved in tissue healing and vascular processes and recruits leukocytes to sites of inflammation and injury by binding to leukocyte receptors[43].VEGFA is an endothelial fibroblast growth factor that acts as a regulator of angiogenesis by controlling endothelial cell integrity and activity,thereby producing optimal vascular morphology and function, and stimulating arterial endothelial cells and macrophages[44], VEGFA has anti-apoptotic and vascular integrity functions in allergic rhinitis.By KEGG enrichment analysis, pivotal target genes associated with epithelial treatment of allergic rhinitis were identified in HIF-1, Tolllike receptor, T-cell receptor and TNF signaling pathways.HIF-1 receptor signaling pathway-mediated transcriptional regulation is an important component of counteracting hypoxia-related cellular stress, and the organism, when fighting infection, slows down blood circulation and consumes more pro-inflammatory cytokines and antigens oxygen, leading to local hypoxia and activation of HIF.HIF can persist indefinitely and participate in the metabolic response to the activation of nuclear transcription factors, which can then activate IL-4-producing macrophages, enhance the activation of NLRP3 inflammatory vesicles, and enhance the release of inflammatory cytokines such as IL-1 and IL-6[45].In mice with allergic rhinitis,inflammatory cells can infiltrate from the nasal mucosa into the extracellular environment of the airways due to the activation of the HIF-1 signaling pathway[46].The TNF signaling pathway is an important pathway in the inflammatory response, and although it can lead to the death of some virally infected cells, it has no effect on normal cells and can even stimulate their development.The T-cell receptor pathway is an important immune response They bind exogenous peptides leading to the activation of T cells, and when the transmitter is activated, CD4+signals to T cells[47], which in turn play an important role in the regulation of allergic rhinitis, thus the T cell receptor signaling pathway plays an important role in the regulation of allergic rhinitis.Toll-like receptors are type I transmembrane proteins natural immune pattern recognition receptors that recognize pathogen-associated molecular patterns in nature, initiating intracellular signaling pathways and activating innate immune responses[48], and genetic variation in TLR signaling pathway genes drives the progression of inflammatory and allergic diseases[49].From the Toll-like signaling pathway map: IL-6 and TNF-α are two pivotal genes linked to the Toll-like receptor signaling pathway,IL-6 and TNF-α are important inflammatory cytokines that are frequently found in various parts of the body inflammatory response,they enhance the inflammatory response and have been shown to be associated with allergic rhinitis[50].The top 10 pivotal overlapping gene network maps of Fuzi for the treatment of allergic rhinitis were analyzed by the network pharmacology-related databases and the MCC algorithm to include mainly IL-6 and TNF-α.Based on these findings, the main target genes IL-6 and TNF-α were experimentally validated in a subsequent animal experimental study.The experimental results showed that Fuzi could mainly reduce the expression activity of IL-6 and TNF-α by inhibiting the activation of Toll-like signaling pathway in the nasal mucosa of mice, thus ameliorating the inflammatory changes in mice with allergic rhinitis.This study was the first to investigate the function and role of Tolllike receptor signaling pathways (IL-6 and TNF-α) in allergic rhinitis, providing a solid foundation for subsequent studies of Fuzi in the treatment of allergic rhinitis.
In conclusion, this study initially explored the main mechanisms of action of Fuzi in the treatment of allergic rhinitis.We explored the possible therapeutic targets and signaling pathways of Fuzi for the treatment of allergic rhinitis on the basis of network pharmacology,providing a scientific basis for the precise treatment of allergic rhinitis with Fuzi, and laying the foundation for the inheritance and innovation of Fuzi.
Note on the contribution of all authors: Zhonglin Mou conceived and designed the study and experiments.Lin Li was responsible for data collection and processing and article writing, and Shun Ding,Zheng-Yang Xu, Jing-Ren Yan, and Qimeng Zhang were responsible for the experimental ideas and the final article revision.All authors have read and approved the final manuscript.
Disclaimer: All authors have no conflict of interest in this article.
猜你喜欢
杂志排行
Journal of Hainan Medical College的其它文章
- Progress in diagnosis and treatment of invasive aspergillosis after liver transplantation
- An overview of the mechanism of acupuncture and moxibustion in treating depression comorbidities insomnia
- The significance of SREBP1 expression in HBV-related hepatocellular carcinoma
- Efficacy and safety of Revlimid combined with Rituximab in the treatment of follicular lymphoma: A meta-analysis
- Effect of acupuncture on acupoint "Yingxiang-Hegu" on Th1, Th2 cytokines and T-bet/GATA-3 of allergic rhinitis rats
- Study on regulating mechanisms of oxocrebanine obtained from Stephania hainanensis H.S.Lo et Y.Tsoong on microtubule sites and tubulin in human breast cancer MCF-7 cells
