A simple and purification-free nucleic acid extraction method for rapid diagnosis of malaria
2023-11-17LeePhoneYouthZenMengYeeLaiMohdHafiziAbdulHamidJenarunJelipRoseNaniMudinNoorHafizanBtMatSallehNettyDarwinaDawamNirzayannaEyanAbdulAjakMohdAshrinAfiqBinZainudinYeeLingLau
Lee Phone Youth Zen ,Meng Yee Lai ,Mohd Hafizi Abdul Hamid ,Jenarun Jelip ,Rose Nani Mudin ,Noor Hafizan Bt Mat Salleh ,Netty Darwina Dawam,Nirzayanna Eyan Abdul Ajak,Mohd Ashrin Afiq Bin Zainudin,Yee Ling Lau✉
1Department of Parasitology, Faculty of Medicine, Universiti Malaya, 50603 Kuala Lumpur, Malaysia
2Vector Borne Disease Sector, Ministry of Health, Putrajaya, Malaysia
3Lipis District Health Office, Pahang, Malaysia
4State Health Department, Negeri Sembilan, Malaysia
5Microbiology Sector, Hospital Kuala Kubu Bharu, Selangor, Malaysia
In a point of care setting for the diagnosis of malaria,DNA extraction using conventional methods are time-consuming and complicated.Therefore,in this study we aim to utilize a simple nucleic acid extraction method to directly extract DNA from blood.This would in turn reduce the time,cost and equipment needed to perform DNA extraction.This method is then coupled with LAMP assay for rapid diagnosis of malaria.
We obtained 77 malaria samples,of which 36 werePlasmodium(P.)knowlesi,10P.vivax,10P.falciparum,1P.malariaeand 20 healthy blood samples from district hospitals from Selangor,Kelantan,Negeri Sembilan,Pahang,and Perak,from 2019 to 2021.All malaria samples tested by LAMP or nested PCR were collected prior to antimalarial treatment.All malaria samples collected were confirmed by microscope at the hospital and cross-checked by Medical Laboratory Technician at the District Health Office.The samples were confirmed by microscopic examination and nested PCR as described by Snounouet al[1] and Imwonget al[2].
The LAMP assay and primers were adapted from Lauet al[3].The extraction method and buffers were adapted from Zouet al[4] with minor modifications.Blood samples of 60 μL,and 240 μL of lysis buffer [800 mM guanidine hydrochloride,50 mM Tris (pH 8),0.5%Triton™X-100,1% Tween-20,40 μg/mL Proteinase K] was used.The tube consisting of blood and lysis buffer mixture was constantly inverted for until homogenous.A 6 mm diameter Whatman grade 1 qualitative filter paper was inserted into the tube was mixed constantly for 1 minute.The filter paper was then removed from the blood lysis mixture and washed in 1 mL of washing buffer[10 mM Tris (pH 8.0),0.1% Tween-20].The filter paper was then mixed constantly in the washing buffer for 1 minute.Following that,the filter paper was removed and dipped 5 times into the PCR tube consisting of the LAMP reaction before removing the filter paper.The LAMP assay was incubated in a Loopamp Real time turbidimeter LA 500 (Eiken Chemical Co.,Ltd.,Japan) at 65 ℃ for 60 minutes and inactivated at 80 ℃ for 2 minutes.
Limit of detection of the method was performed by usingP.knowlesistrain A1H1 obtained from the Department of Parasitology,Faculty of Medicine,Universiti Malaya.P.knowlesistrain A1H1 culture blood was ten-fold serially diluted to parasitemia of 1%,0.1%,0.01%,0.001% and 0.000 1% respectively with healthy blood,and was tested with the above method in triplicates.
The clinical sensitivity and specificity of LAMP assay was determined using microscopy as the reference standard methods.Sensitivity was calculated as (number of true positives)/(number of true positives+number of false negatives),and specificity was calculated as (number of true negatives)/(number of true negatives+number of false positives).
The DNA extraction method coupled with LAMP has a detection limit of 0.001% parasitemia (5 parasites/μL of blood) (Figure 1).A highly trained technician using microscopy can reliably detect as few as 50 parasites/μL of blood,while the published limit of detection of laboratory PCR methods is 0.5 to 5 parasites/μL[5].When conducting the limit of detection,all samples with parasitemia higher than 0.001% was able to amplify successfully,whereas there was a 1/3 positive amplification at 0.000 1% parasitemia.Clinical sensitivity of LAMP was compared with the results from conventional nested PCR and microscopy.A total of 77 samples were diagnosed using both nested PCR and microscopy.Within the 77 samples diagnosed using both nested PCR and microscopy,there wereP.knowlesi(n=36),P.falciparum(n=10),P.vivax(n=10),P.malariae(n=1) and 20 negativePlasmodiumsamples.Among 36P.knowlesi,3 were not detected using the alternative DNA extraction method coupled with LAMP.This method has successfully amplified allP.falciparum(n=10),P.vivax(n=10) andP.malariae(n=1) positive samples.The 20 healthy blood samples did not show any amplification.With this,the DNA extraction method coupled with LAMP showed 94.7% sensitivity and 100% specificity when compared with microscopy.
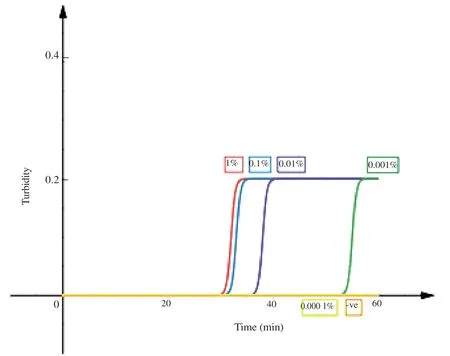
Figure 1.Limit of detection of the DNA extraction method coupled with loop mediated isothermal amplification.A graph was plotted based on the results obtained from the Loopamp Real time turbidimeter LA 500 (Eiken Chemical Co.,Ltd.,Japan).The X-axis represents time whereas,the Y-axis represents the turbidity.The labels are as follows,1%: 1% parasitemia Plasmodium knowlesi A1H1 culture;0.1%: 0.1% parasitemia Plasmodium knowlesi A1H1 culture;0.01%: 0.01% parasitemia Plasmodium knowlesi A1H1 culture;0.001%: 0.001% parasitemia Plasmodium knowlesi A1H1 culture;0.000 1%: 0.000 1%parasitemia Plasmodium knowlesi A1H1 culture;-ve: double distilled H2O (no target control).
The alternative DNA extraction method coupled with LAMP has a clinical sensitivity of 94.7% and a 100% clinical specificity when compared with microscopy.Three samples failed to amplify by LAMP may be due to degradation of DNA.In order to confirm the degradation of DNA,these three samples were subjected to nested PCR and no amplification was observed.These samples were kept at -20 ℃ for more than a year.Short-term storage at room temperature,4 ℃ and -20 ℃ will affect the yield of DNA greatly[6].Extracting genomic DNA by conventional methods such as isopropanol precipitation,formamide lysate method,nonorganic solvent extraction,and glass particle adsorption is a timeconsuming process while,the phenol and chloroform method uses toxic reagents that are not fit for on field diagnostics.Majority of commercial DNA extraction kits require multiple liquid handling steps[7] which is not suitable for point of care.Another approach of the DNA extraction method is by using Fitzco/Flinder Technology Agreement (FTA) paper,a cellulose based DNA extraction method.FTA paper is an absorbent cellulose-based paper that has been treated with a proprietary mix of chemicals that allows good preservation and storage capabilities.However,FTA based DNA extraction method is still considered costly when compared to alternatives that use untreated filter paper.Based on the Whatman price catalog[8],a single use FTA blood kit costs 10.48 USD,whereas one 70 mm diameter Whatman grade 1 qualitative filter paper for the proposed extraction method only costs 0.11 USD for multiple samples.A paper published by Zouet al.[4] found that cellulose based filter paper can be used to rapidly bind nucleic acids,retain them during a short washing step to remove contaminants,followed by elution directly into the amplification reaction.They then adapted the filter paper into a dipstick and was able to extract nucleic acids from a wide range of biological samples in less than 30 seconds without the need of any specialised equipment[4].We adapted this method in this study for the diagnosis of malaria.A 6-mm diameter filter paper was used instead so that the bigger surface area would aid the binding of the DNA.The volume of the washing buffer was increased to 1 mL instead of 200 μL,as we found that a larger volume helped the diffusion of contaminants from the filter paper.The filter paper was then dipped into a LAMP assay allowingPlasmodiumparasite DNA templates to be present in the assay.
In conclusion,the method presented here allows amplification ofPlasmodiumDNA in a time and cost-effective manner.Diagnosis of malaria is more accessible and affordable with this method,so it will be useful in resource limited areas.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Medical Ethics Committee of UMMC (MEC reference No.817.18 and 908.11) and National Medical Research Registry (reference No.NMRR-12-1105-13079).
Acknowledgements
We sincerely thank the local health officers and the hospital staff from Hospital Kuala Lipis and Klinik Kesihatan Betau (Pahang);Hospital Sungai Siput,Hospital Permaisuri Bainun,Hospital Kuala Kangsar (Perak);Vector lab (Negeri Sembilan);Hospital Gua Musang,Hospital Sultan Ismail Petra and Hospital Jeli (Kelantan);and Hospital Kuala Kubu Bahru (Selangor) who had helped in sampling.
Funding
This study was supported by Long Term Research Grant Scheme(LRGS),LRGS/1/2018/UM/01/1/4 from the Ministry of Higher Education,Malaysia.
Authors’contributions
LPYZ and YLL conceptualized the study.LPYZ curated the data,formally analysed,investigated,validated and wrote the original draft.YLL acquired funding and administered the project.YLL and MYL supervised the project.MHAH,JJ,RNM,NHMS,NDD,NEAA and MAABZ contributed resources.All the authors were involved in the writing review and editing.
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- Elimination of lymphatic filariasis: Where do we stand so far?
- Attitudes and understanding of complementary and alternative medicine in cancer care: An exploratory study of patients’ perspectives in Karachi,Pakistan
- Public satisfaction with the quality of First Health Facility Services in Indonesia: Does sociodemographic matter?
- Efficacy and safety of Lianhuaqingwen capsules in high-risk common type COVID-19 pneumonia: A multicenter retrospective study
- Type 2 reaction associated sensorineural hearing loss in a drug resistant lepromatous leprosy patient: A case report
