Protein Containing the GGDEF Domain Affects Motility and Biofilm Formation in Vibrio cholerae and is Negatively Regulated by Fur and HapR*
2023-11-14GAOHeMALiZhiQINQinCUIYaoMAXiaoHanZHANGYiQuanandKANBiao
GAO He, MA Li Zhi, QIN Qin, CUI Yao, MA Xiao Han, ZHANG Yi Quan, and KAN Biao,#
1. State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China; 2. Medical Risk Management Section, Third Medical Centre, Chinese PLA (People’s Liberation Army) General Hospital, Beijing 100039, China; 3. Department of Clinical Laboratory, Affiliated Nantong Hospital 3 of Nantong University, Nantong Third People's Hospital, Nantong 226006, Jiangsu, China
Abstract
Key words: Vibrio cholera; Cyclic di-GMP; VCA0560; Fur; HapR
INTRODUCTION
Vibrio choleraenaturally lives in aquatic environments and causes deadly cholera in humans[1].V.choleraebiofilms play significant roles in virulence, persistence,transmission, and survival in adverse environments[2-5],and their formation requires specific structures,including flagella, exopolysaccharide, and pili[5].Numerous factors, such as VpsR[6,7], VpsT[6,7], VxrB[8],H-NS[9], CRP[10], CarR[11], PhoB[12], Fur[13], quorum sensing (QS)[14,15], and cyclic diguanylate (c-di-GMP)[16], are required for biofilm regulation inV.cholerae.
Cyclic di-GMP (c-di-GMP) is diffusely applied by bacteria to modulate multiple behaviors, including motility, biofilm formation, virulence determinant production, DNA repair, and cell shape[16-18].Increased c-di-GMP levels in bacterial cells induce biofilm formation, and decreased c-di-GMP concentrations induce a motile lifestyle[16,19].c-di-GMP is catalyzed by diguanylate cyclase (DGC)harboring the GGDEF domain and is degraded by phosphodiesterase harboring the EAL or HD-GYP domain[16].More than 50 genes encode GGDEFand/or EAL-type proteins inV.cholerae[20], and about 50% of these genes contribute to c-di-GMP metabolism[10,19,21-33].However, the c-di-GMP metabolic pool inV.choleraewarrants further investigation.
The VCA0560 gene encodes a GGDEF-type protein inV.cholerae[34].VCA0560 overexpression reduces motility[35]but only slightly alters biofilm formation[36].Whether the VCA0560 protein affects the intracellular c-di-GMP pool needs to be further investigated.Here, the data showed that VCA0560 gene deletion does not affect the c-di-GMP synthesis, biofilm formation, and motility.Meanwhile, VCA0560 overexpression enhances c-di-GMP production and biofilm formation but reduces swimming motility.In addition, the transcription of the VCA0560 gene is directly repressed by the master quorum sensing (QS) regulator HapR[37]and the ferric uptake regulator (Fur)[38].In summary,overexpressed VCA0560 acts as an active DGC, and its transcription is repressed by Fur and HapR.
MATERIALS AND METHODS
Bacterial Strains
V.choleraeEl Tor C7258 was applied as the wildtype (WT) strain.NonpolarhapRandfursingle-gene mutants (ΔhapRand Δfur) and the complementary mutants ΔhapR/pBAD24-hapR(C-ΔhapR) and Δfur/pBAD24-fur(C-Δfur) were constructed as previously described[39].The VCA0560 gene mutant(ΔVCA0560) was constructed from WT using the suicide plasmid pWM91 by the allelic exchange as previously described[40].The coding region of the VCA0560 gene was cloned into the pBAD24 vector containing an arabinose-inducing pBAD promoter to construct the overexpression plasmid pBAD24-VCA0560[41].The recombinant pBAD24-VCA0560 was transferred into WT to construct the VCA0560 overexpression strain WT/pBAD24-VCA0560, which produced about 3.5 times as many VCA0560 mRNA levels as that of the WT/pBAD24 strain (data not shown).The pBAD24 vector was also introduced into WT, ΔhapR, and Δfurto prepare controls.All primers used are listed in Table 1.
Growth Conditions
Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, and 1% NaCl) was applied forV.choleraecultivation[39].Overnight bacterial cultures were diluted at 1:50 into 5 mL of fresh LB broth and grown at 30 °C with shaking at 200 rpm to an OD600value of 1.0.The resultant cultures were diluted 1:100 into 5 mL of fresh LB broth for the third-round growth.The bacterial cells were harvested at an OD600value of about 0.6.Whenever required, antibiotics were used in the following concentrations: 100 mg/mL ampicillin,5 mg/mL chloramphenicol, and 0.1% arabinose.

Table 1.Oligonucleotide primers used in this study
Colony Morphological Characterization
Colony morphology was characterized as previously described[42].In brief, 2 µL of overnight bacterial culture was spotted onto the LB agar and then statically incubated at 30 °C for 48 h.
Crystal Violet (CV) Staining
CV staining was performed as previously described[42].The third round of cultures was diluted 50-fold into 2 mL of fresh LB broth in glass tubes and then allowed to grow at 30 °C with shaking at 100 rpm for 48 h.The surface-attached cells were stained with 0.1% CV.The bound dye was dissolved with dimethylsulfoxide (DMSO), and the OD570values were measured for each strain as the index of CV staining.
Quantification of c-di-GMP
Intracellular c-di-GMP levels were detected as previously described[13,43].Bacterial cells were harvested at OD600= 0.6, washed twice with ice-cold phosphate buffered solution (PBS), resuspended in 2 mL of ice-cold PBS, incubated at 100 °C for 5 min,and sonicated for 15 min (power 100%, frequency 37 kHz) in an ice-water bath.The supernatant was collected, and the pellet was resuspended in 2 mL of ice-cold PBS and re-extracted twice.The intracellular c-di-GMP levels were determined with a c-di-GMP ELISA kit (Mskbio, Beijing, China).Total protein concentration in the supernatant was also determined using a Pierce BCA Protein Assay kit(ThermoFisher Scientific, USA) in accordance with the manufacturer’s instructions.c-di-GMP concentration was expressed as pmol/mg protein.
Swimming Motility Assay
Swimming motility was assayed as previously described[44].In brief, 2 µL of the third round of cell cultures were inoculated into semisolid swim plates(1% tryptone, 0.5% yeast extract, 1% NaCl, and 0.35% bacterial agar), and the diameter of the swimming areas was measured after incubation at 30 °C for about 10 h.
Luminescence Assay
A luminescence assay was performed as previously described[45].The regulatory DNA region of VCA0560 was cloned into the pBBRlux vector harboring a promoterlessluxCDABEreporter gene.The recombinant plasmid was transferred into WT and regulatory gene mutants (ΔhapRand Δfur) to measureluxactivity in each strain.Luminescence was determined using an Infinite® 200 Pro NanoQuant (Tecan, Switzerland), andluxactivity was calculated as light units/OD600.
Preparation of His-Tagged Fur and HapR (His-Fur and His-HapR)
The coding regions ofhapRandfurwere separately cloned into the pET28a (Novagen, USA) to express His-HapR and His-Fur usingEscherichia coliBL21λDE3 cells[46].His-HapR and His-Fur were prepared as previously described[47,48].The dialyzed proteins were concentrated to approximately 0.5 mg/mL, and their purity was analyzed by SDS-10% PAGE.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as previously described[48].The 5ʹ-ends of the regulatory DNA region of the VCA0560 gene were labeled with [γ-32P] ATP and T4 polynucleotide kinase.DNA binding was performed in a 10 µL reaction volume containing binding buffer(1 mmol/L MgCl2, 100 µmol/L MnCl2or 0.5 mmol/L EDTA, 0.5 mmol/L DTT, 50 mmol/L NaCl, 10 mmol/L Tris-HCl/pH 7.5, and 10 mg/mL salmon sperm DNA),labeled DNA probe, and increasing amounts of purified His-Fur or His-HapR.After incubation at room temperature for 20 min, the products were loaded onto a native 4% (w/v) polyacrylamide gel and electrophoresed in 1.0× TB or 0.5× TBE buffer for approximately 60 min at 150 V.Radioactive species were detected by autoradiography after exposure to Fuji Medical X-ray film at -20 °C.Three controls were included in each EMSA experiment:1) a cold probe as a specific DNA competitor (the same promoter DNA region unlabeled), 2) a negative probe as a nonspecific DNA competitor (the unlabeled coding region of the 16S rRNA gene), and 3) a nonspecific protein competitor (rabbit anti-F1-protein polyclonal antibodies).
DNase I Footprinting
DNase I footprinting was performed as previously described[49-51].The results were analyzed using an ABI 3500XL DNA Genetic analyzer with GeneMarker software 2.2.
Statistical Methods
Colony morphological characterization, CV staining, EMSA, and DNase I footprinting assays were performed at least twice with similar results.Luminescence assay and c-di-GMP quantification were performed three times, and the values were expressed as mean ± standard deviation (SD).Paired Student’st-tests were applied to calculate statistical significance.Pvalues less than 0.01 were considered significant.
RESULTS
Growth of V. cholerae Strains
The growth curves of WT/pBAD24,ΔVCA0560/pBAD24, and WT/pBAD24-VCA0560 grown at 30 °C in LB broth were recorded with 1 h intervals to determine the effect of VCA0560 on the growth ofV.cholerae.As shown in Figure 1,indistinguishable growth rates were observed for WT/pBAD24 and WT/pBAD24-VCA0560, indicating that VCA0560 overexpression did not affect the growth ofV.cholerae.However, the ΔVCA0560/pBAD24 strain showed a slight growth defect relative to the WT/pBAD24 and WT/pBAD24-VCA0560 strains, suggesting that VCA0560 might be required for the growth ofV.cholerae.
VCA0560 Overexpression Induced V. cholerae Biofilm Formation
Colony morphological characterization and CV staining were performed to compare colony morphology and biofilm quantity among WT/pBAD24, ΔVCA0560/pBAD24, and WT/pBAD24-VCA0560.As shown in Figure 2A, WT/pBAD24 and ΔVCA0560/pBAD24 produced smooth colonies on the LB plates, and WT/pBAD24-VCA0560 formed

Figure 2.VCA0560 overexpression induced biofilm formation by V. cholerae.(A) Colony morphological characterization.In brief, 2 µL of overnight bacterial culture was spotted on the LB agar supplemented with 100 mg/mL ampicillin and 0.1% arabinose and then statically incubated at 30 °C for 48 h.(B) Crystal violet (CV) staining.Bacterial cells were inoculated into 2 mL of fresh LB broth supplemented with 100 mg/mL ampicillin and 0.1% arabinose in glass tubes and allowed to grow at 30 °C with shaking at 100 rpm for 48 h.The surface-attached cells were stained with 0.1% CV.The bound CV was dissolved with dimethylsulfoxide (DMSO), and the OD570 values were measured as the index of CV staining.NS represents no significant difference.
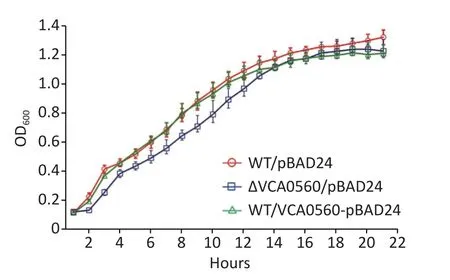
Figure 1.Growth curves of V. cholerae strains.The overnight growth cultures of WT/pBAD24,ΔVCA0560/pBAD24, and WT/pBAD24-VCA0560 were diluted at 1:100 into 20 mL of LB broth supplemented with 100 mg/mL ampicillin and 0.1% arabinose, mixed well, and then divided into a microwell plate with 300 µL in each well.V. cholerae strains were grown at 30 °C with shaking at 200 rpm, and the OD600 values were monitored at 1 h intervals.The experiment was conducted twice, with five replicates per trial for each strain.
wrinkled colonies.CV staining assay showed that WT/pBAD24-VCA0560 produced significantly more normalized CV staining than WT/pBAD24 and ΔVCA0560/pBAD24, and ΔVCA0560/pBAD24 showed similar CV staining result with WT/pBAD24(Figure 2B).These results indicated that VCA0560 gene deletion had no regulatory activity on biofilm formation, but overexpression of the protein did induce biofilm formation byV.cholerae.
VCA0560 Overexpression Inhibited V. cholerae Swimming Motility
The swimming motility assay showed that the swimming capacity of WT/pBAD24-VCA0560 was significantly inhibited relative to those of WT/pBAD24 and ΔVCA0560/ pBAD24.ΔVCA0560/pBAD24 and WT/pBAD24 manifested a similar capacity for swimming motility (Figure 3).These results suggested that VCA0560 overexpression inhibitedV.choleraeswimming motility.
Overexpression of VCA0650 Induced c-di-GMP Production

Figure 3.Regulation of swimming motility by VCA0560 protein in V. cholerae.In brief, 2 µL of bacterial cells were inoculated into semisolid swim plates (1% tryptone, 0.5%yeast extract, 1% NaCl, and 0.35% bacterial agar) supplemented with 100 mg/mL ampicillin and 0.1% arabinose.The diameter of the swimming areas was measured after incubation at 30 °C for 10 h.
Elevated c-di-GMP levels in bacterial cells lead to decreased motility but increased biofilm formation[5].Thus, we further detected whether VCA0560 regulates c-di-GMP production inV.cholerae.The concentration of c-di-GMP in WT/pBAD24-VCA0560 was higher than that in WT/pBAD24 and ΔVCA0560/pBAD24.Meanwhile, ΔVCA0560/pBAD24 had similar c-di-GMP levels to WT/pBAD24 (Figure 4).These results indicated that VCA0560 mutation did not influence c-di-GMP production, but VCA0560 overexpression induced c-di-GMP synthesis inV.cholerae.
Fur Directly Repressed VCA0560 Transcription
A Fur consensus-like sequence AATAAGCATA GTTTTCATC with a weighted score of 8.1 located from -126 bp to -108 bp was detected within the upstream of the VCA0560 gene[38,52], suggesting that VCA0560 transcription might be directly regulated by Fur inV.cholerae.In a previous study, Fur has been demonstrated to repress c-di-GMP production and biofilm formation byV.choleraeindependent of iron[13].Thus, the bacterium was cultured in LB broth and then harvested to investigate the Fur-dependent transcription of VCA0560.As shown in Figure 5A, the luminescence activity under the direct action of the VCA0560 promoter was higher in Δfurthan in WT across the growth periods.EMSA results indicated that His-Fur dose-dependently bound to the 581 bp upstream of the VCA0560 gene but was incapable of binding to the negative control DNA fromrecA(Figure 5B).DNase I footprinting assay revealed that His-Fur protected a single DNA region located from-162 bp to -19 bp upstream of VCA0560 (Figure 5C).Thus, VCA0560 transcription was directly suppressed by Fur.

Figure 4.Intracellular c-di-GMP levels in V.cholerae strains.V. cholerae strains were cultured in LB broth supplemented with 100 mg/mL ampicillin and 0.1% arabinose at 30 °C and harvested at OD600 = 0.6.Intracellular c-di-GMP was extracted by ultrasonication, and its concentration was measured by a c-di-GMP ELISA kit (Mskbio,Beijing, China) and expressed as pmol/mg.NS represents no significant difference.
HapR Directly Repressed VCA0560 Transcription
An MQSR consensus-like sequence TAATTGTA GAATTATCATTA with a weighted score of 8.3 located from -46 bp to -27 bp was detected within the upstream of the VCA0560 gene[48], suggesting that VCA0560 transcription might be directly regulated by HapR.As shown in Figure 6A, VCA0560 expressed was enhanced significantly in ΔhapRcompared with that in WT across the growth periods.EMSA results demonstrated that His-HapR dose-dependently bound to the regulatory DNA region of the VCA0560 gene but was incapable of binding to the negative control DNA fragment fromrecA(Figure 6B).DNase I footprinting assay results showed that His-HapR bound one DNA region located from -59 bp to -11 bp upstream of the coding region of VCA0560(Figure 6C).Thus, VCA0560 transcription was directly inhibited by HapR.
DISCUSSION
In this work, we showed that the overexpression of the VCA0560 gene, which encodes a GGDEF-type protein inV.cholerae, induced biofilm formation and c-di-GMP production but inhibited the motility ofV.cholerae.Meanwhile, the deletion of the VCA0560 gene had no influence on c-di-GMP synthesis, biofilm formation, and motility (Figures 2–4).These results suggested that the VCA0560 protein acts as an active DGC inV.cholerae, but its activity is low.A previous study also showed that VCA0560 overexpression seems to have only a weakly positive effect on the biofilm formation byV.cholerae[36].The deletion of a DGC gene does not alter the phenotype changes because more than 30 GGDEF-containing proteins with a high level of functional redundancy can be found inV.cholerae[20].The VCA0560 protein is perhaps not the main enzyme that modulates c-di-GMP synthesis under the growth conditions in the current work.VCA0560 expression is significantly induced in the early stage than in the late stage of cholera infection[53].V.choleraeforms biofilms almost immediately after it adheres to the intestinal cells[2,3].Thus, the VCA0560 protein might play some roles in the early stage ofV.choleraeinfection by promoting c-di-GMP synthesis and biofilm formation.Further investigations are warranted to verify this hypothesis.

Figure 6.HapR regulated the transcription of the VCA0560 gene.The first base of the start codon of VCA0650 was defined as +1.Luminescence assay (A), EMSA (B), and DNase I footprinting (C) were performed as described in Figure 5.
A Fur consensus-like sequence and an MQSR consensus-like sequence were detected in the regulatory DNA region of the VCA0560 gene.Thus,we further investigated the transcriptional regulation of the VCA0560 gene by Fur and HapR.The data showed that VCA0560 transcription was directly repressed by HapR and Fur (Figures 5 and 6).In the presence and absence of HapR and Fur,the promoter activity of the VCA0560 gene increased with cell density until OD600= 0.6 and then decreased over time (Figures 5A and 6A).Thus, VCA0560 might be repressed strongly by some other unknown regulators operating at low and high cell densities.In addition, primer extension[13,39]and 5ʹ-RACE[13]did not detect any transcription starts for VCA0560 (data not shown).Only one transcription start located at 124 bp upstream of the VCA0560 gene was predicted using the online tool SoftBerry (http://linux1.softberry.com/) (Figure 7).The putative -10 element was well matched to the -10 consensus,but the putative -35 element possessed three bases mismatch with the consensus (Figure 7),suggesting that the promoter is relatively weak.The Fur site overlapped with the putative -10 and-35 elements and the predicted transcription start, indicating that Fur represses VCA0560 transcription possibly via its direct interference with RNA polymerase (RNAP) function.In contrast,the HapR site was located downstream of the predicted transcription start.Thus, HapR represses VCA0560 transcription, possibly by blocking RNAP elongation.The HapR site almost entirely overlapped with the Fur site (Figure 7).Whether HapR cooperates with Fur to repress VCA0560 transcription or not warrants further investigation.
HapR acts as a negative regulator of biofilm formation, intracellular c-di-GMP synthesis, and many other gene loci inV.cholerae[15,54-56].In addition, HapR regulates the transcription of 14 genes involved in c-di-GMP metabolism inV.cholerae, but the VCA0560 gene is not included[15].The above data were obtained in the genetic background ofluxOmutant[15].LuxO is a central quorum regulator that controls the QS circuit and many other genes[57,58].Therefore, the discrepancies between previous and present data may be mainly due to the bacterial genetic background.Moreover,Fur acts as a repressor of biofilm formation and c-di-GMP synthesis inV.cholerae[13], and its transcription is directly repressed by HapR[39].The current study showed that Fur and HapR directly repressed the transcription of VCA0560 encoding a GGDEF-type protein that acts as an active DGC inV.cholerae.Thus, Fur and HapR compose a regulatory circuit to tightly regulate biofilm formation at multiple genes and levels.
In conclusion, VCA0560 overexpression significantly enhances biofilm formation and intracellular c-di-GMP levels but reduces the motility capacity ofV.cholerae.Fur and HapR bind to the regulatory DNA region of the VCA0560 gene to repress its transcription.Therefore, overexpressed VCA0560 acts as an active DGC inV.cholerae, and its transcription is repressed by Fur and HapR.
Received: November 10, 2022;
Accepted: January 26, 2023
杂志排行
Biomedical and Environmental Sciences的其它文章
- Treatment Outcomes in COVlD-19 Patients with Brucellosis:Case Series in Heilongjiang and Systematic Review of Literature*
- Value of Pretreatment lnflammation-nutrition Score to Predict Non-response to Neoadjuvant Chemotherapy in Locally Advanced Rectal Cancer*
- A Comprehensive View on the Progress of Organoid Research with an Emphasis on its Relevance to Disease Characterization
- Altered Expression of COMP, Collagen 1, and MMP9 in Pelvic Organ Prolapse*
- Synthesis of TiO2 Nanospikes for Dual Antibacterial Activity*
- Aurora A Kinase Plays a Key Role in Mitosis Skip during Senescence lnduced by lonizing Radiation*
