Cloning and Bioinformatics Analysis of vscB Gene of T3SS Chaperone of Vibrio alginolyticus
2023-05-16HongweiZHENGLiangchuanCHENHaiyunFENGYunshengCHANGYuDINGWeijieZHANGHuanyingPANGNaWANG
Hongwei ZHENG, Liangchuan CHEN, Haiyun FENG, Yunsheng CHANG, Yu DING, Weijie ZHANG, Huanying PANG*, Na WANG
1. Shenzhen Institute of Guangdong Ocean University, Shenzhen 510000, China; 2. Fisheries College, Guangdong Ocean University, Zhanjiang 524025, China; 3. Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture &Key Laboratory of Control for Diseases of Aquatic Economic Animals of Guangdong Higher Education Institutes, Zhanjiang 524025, China; 4. Chinese Academy of Inspection and Quarantine, Beijing 100176, China
Abstract [Objectives] To clone and analyze the vscB gene of Vibrio alginolyticus HY9901 by bioinformatics. [Methods] A pair of specific primers were designed according to the vscB gene sequence of Vibrio alginolyticus HY9901. The full length of the primers was cloned by PCR and analyzed by bioinformatics. [Results] The vscB gene was 429 bp long, encoding 142 amino acids, with a theoretical molecular weight of 16.4 kDa and a pI value of 5.48. Amino acid sequence analysis of VscB showed that VscB was not a secretory protein, without signal peptide and transmembrane region, and there were protein kinase C phosphorylation site and casein kinase II phosphorylation site in the sequence. Homologous comparison of amino acid sequences showed that VscB of V. alginolyticus had the highest protein similarity with Vibrio Parahaemolyticus, reaching 91%. Phylogenetic tree analysis showed that the corresponding proteins of V. alginolyticus VscB, Vibrio Parahaemolyticus and Vibrio diabolicus were clustered in the same subfamily. Functional domain analysis showed that it had CesT family domain. Tertiary structure prediction showed that there were 3 α-helices and 5 β-turns in VscB protein. [Conclusions] This study provided a theoretical basis for further study on the function of chaperone of V. alginolyticus.
Key words Vibrio alginolyticus, Gene cloning, vscB, Bioinformatics analysis
1 Introduction
Vibrioalginolyticus, as one of the main pathogens of marine aquatic economic animals, has caused serious harm to aquaculture industry[1]. As a virulence factor ofV.alginolyticus, Type III secretion system (T3SS) is closely related to its pathogenicity. T3SS can be divided into five types of proteins according to their functions, which are regulatory proteins, effector proteins, device proteins, molecular chaperones and regulatory proteins[2]. Different proteins have different effects on the host. Although there have been many reports on the mechanism of T3SS effector protein[3], there are still few studies on the effect of T3SS chaperone, which is necessary for maintaining the stability of effector protein in bacterial cytoplasm[4]. Molecular chaperones also assist in the secretion or transport of T3SS effector proteins[5], and some can also enter host cells[6-7]. VscB protein is an important component of T3SS, which is supposed to play an important role in host invasion. However, the function of chaperone VscB ofV.alginolyticusT3SS is still unclear. Therefore, cloning and bioinformatics analysis ofvscBgene ofV.alginolyticusHY9901 will provide theoretical basis for further study on the function of chaperone ofV.alginolyticus.
2 Materials and methods
2.1 Materials
2.1.1Strains
V.alginolyticusHY9901, isolated from diseased red snapper in Zhanjiang Port, Guangdong Province[8], was stored in the ultra-low temperature refrigerator at -80 ℃ in this laboratory; the receptor strainE.coli(DH5α) was preserved in this laboratory; the pMD18-T (Amp+) cloning vector was purchased from Takara Biotechnology.
2.1.2Reagents. TIANamp bacteria DNA kit and DNA gel recovery kit, kit (Tiangen Biotech); Ex Taq DNA polymerase (Takara Biotechnology); primer synthesis and sequencing (Shanghai Sangon Co., Ltd.).
2.1.3Instruments. Electrophoresis instrument and PCR instrument (Bio-Rad Life Medicine Co., Ltd.); ultrapure water instrument (Beijing Rightleder Water Treatment Equipment Co., Ltd.); refrigeration high-speed centrifuge: (Eppendorf Life Sciences Co., Ltd.); HVE-50 series autoclave: (Hirayama); gel imaging equipment: (Priotein Simple); ultraviolet spectrophotometer: (Shimadzu); ultra-low temperature refrigerator: (Thermo Fisher).
2.2 Methods
2.2.1Genome extraction ofV.alginolyticusstrain HY9901.V.alginolyticuswas removed from the ultra-low temperature refrigerator and marked on the TSA plate; single colonies were selected and inoculated in TSB medium according to the ratio of 1∶100; it was cultured in a shaking table at 28 ℃ at a rotational speed of 180 r/min for more than 14 h, and a small amount of bacteria liquid was transferred into a centrifuge tube; a high-speed centrifuge was used to centrifuge at 10 000 r/min for 2-3 min, the supernatant was discarded andV.alginolyticuswas collected; according to the bacterial genome extraction kit, the total DNA ofV.alginolyticuswas extracted; it was refrigerated at -20 ℃ for later use.
2.2.2Cloning ofvscBandvscDgenes. According to the whole gene sequence ofV.alginolyticusin GenBank (Accession: GU074526), a pair of primers were designed: the forward primer F1 ofvscBwas ATGTTAGATAAGATGATGAAATC; the reverse primer R1 was TCATGGTAACCACACTGTATG. PCR reaction conditions:vscB—pre-denaturation at 95 ℃ for 5 min; denaturation at 95 ℃ for 30 s, annealing at 60 ℃ for 30 s and extension at 68 ℃ for 30 s, a total of 30 cycles; extension at 72 ℃ for 10 min. PCR products were detected by 1% agarose gel electrophoresis, and then glue recycling kit was used for glue recycling.
2.2.3Ligation and sequencing of the target fragment to the vector.vscBand pMD 18-T vector were ligated overnight at 4 ℃, and the ligated products were transferred into competentE.coliDH5α cells, cultured at 37 ℃, and the positive clones were tested after colony PCR detection.
2.2.4Web sites for bioinformatics analysis[9]. The amino acid sequence, theoretical isoelectric point (pI) and molecular weight of the target protein were predicted online by ExPASy Proteomics Server; SignalP4.1 Server was used to predict whether the target protein contains signal peptide; GeneDoC was used for homologous comparison of amino acid sequences of the target protein; the phylogenetic tree was constructed by Clastal 2.0 and MEGA 6.0; TMHMM Server 2.0 was used to predict whether the target protein contains transmembrane structure; the functional sites in protein amino acid sequence were analyzed by SoftBerry-Psite; the domain of the target protein was predicted by NCBI conserved domain database CDD; SWISS-MODEL was used to predict and construct the tertiary structure of protein online.
3 Results and analysis
3.1 Full-length cloning ofvscBgeneAvscB-specific band with a length of about 400 bp was obtained by PCR reaction (Fig.1). The open reading frame (ORF) ofvscBwas 429 bp and 142 amino acids were encoded.vscBwas submitted to GenBank withvscBlogin number: MG905226.
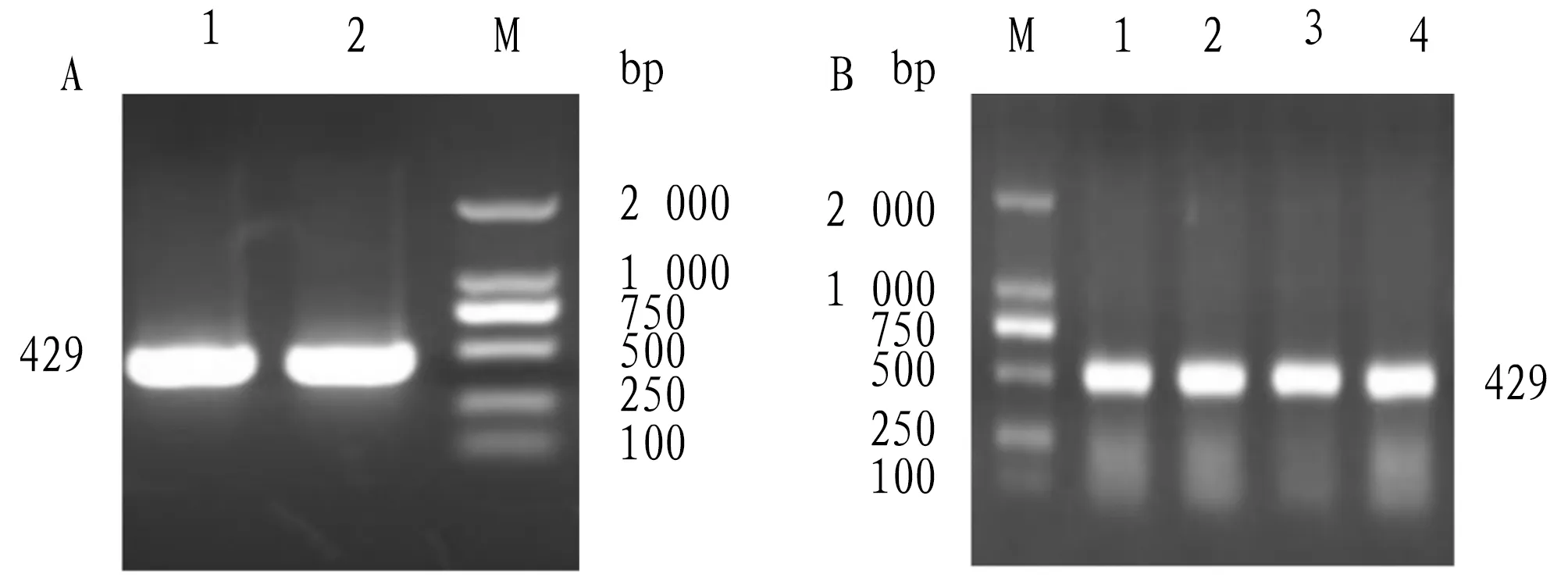
Note: A. PCR amplification of vscB gene; B. Colony PCR identification of vscB positive colonies.Fig.1 Cloning of vscB gene
3.2 Physicochemical properties of VscBVscB protein ofV.alginolyticuswas analyzed by ExPASy. The results showed that the total number of atoms of VscB protein was 2 308, and its molecular structural formula was C726H1157N199O219S7. The theoretical molecular weight was 16.402 kDa and the theoreticalpIvalue was 5.48. The instability coefficient was 38.76, which indicated that the protein was stable; the fat coefficient was 101.62, and the total average hydrophilicity was -0.251. The total number of acidic amino acid residues (Asp+Glu) and basic amino acids (Arg+Lys) was 19 and 14, respectively. The N-end of the protein sequence was methionine (Met). The estimated half-life was 20 h in yeast and 10 h inE.coli. When expressedinvitro, the half-life of mammalian reticular cells was about 30 h.
3.3 VscB sequence analysisSignalP 4.0 Server was used to analyze the N-end signal peptide sequence of VscB protein. The results showed that vscB had no signal peptide, which indicated that VscB was not a secretory protein. The prediction results of TMHMM Server 2.0 showed that VscB had no transmembrane region. SoftBerry-Psite predicted that there were three potential phosphorylation sites of protein kinase C and four phosphorylation sites of casein kinase II in VscB amino acid sequence (Fig.2).

Note: * for terminator, underline for protein kinase C phosphorylation site, gray for tyrosine kinase II phosphorylation site, and wavy line for cAMP and cGMP dependent protein kinase phosphorylation site.Fig.2 Nucleotide of vscB gene and its encoded amino acid sequence
3.4 Homologous and evolutionary analysisClustalX molecular software was used to compare the amino acid sequences of VscB ofV.alginolyticus, and the phylogenetic trees of VscB were constructed by MEGA 6.0. VscB ofV.alginolyticuswas found to have high homology with VscB of other species of Vibrio by BLAST, and VscB ofV.alginolyticusandVibrioParahaemolyticushad the highest similarity, reaching 91%. The results of multi-sequence alignment showed that VscB was relatively conservative in Vibrio (Fig.3). Through phylogenetic tree analysis, the VscB protein ofV.alginolyticusand the corresponding proteins ofVibrioParahaemolyticusand Vibrio diabolicus were clustered in the same subfamily, indicating that they were close in evolution (Fig.4).
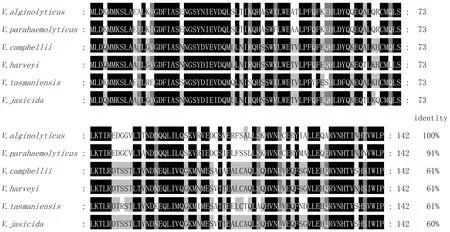
Fig.3 Homologous comparison of deduced amino acid sequences of VscB
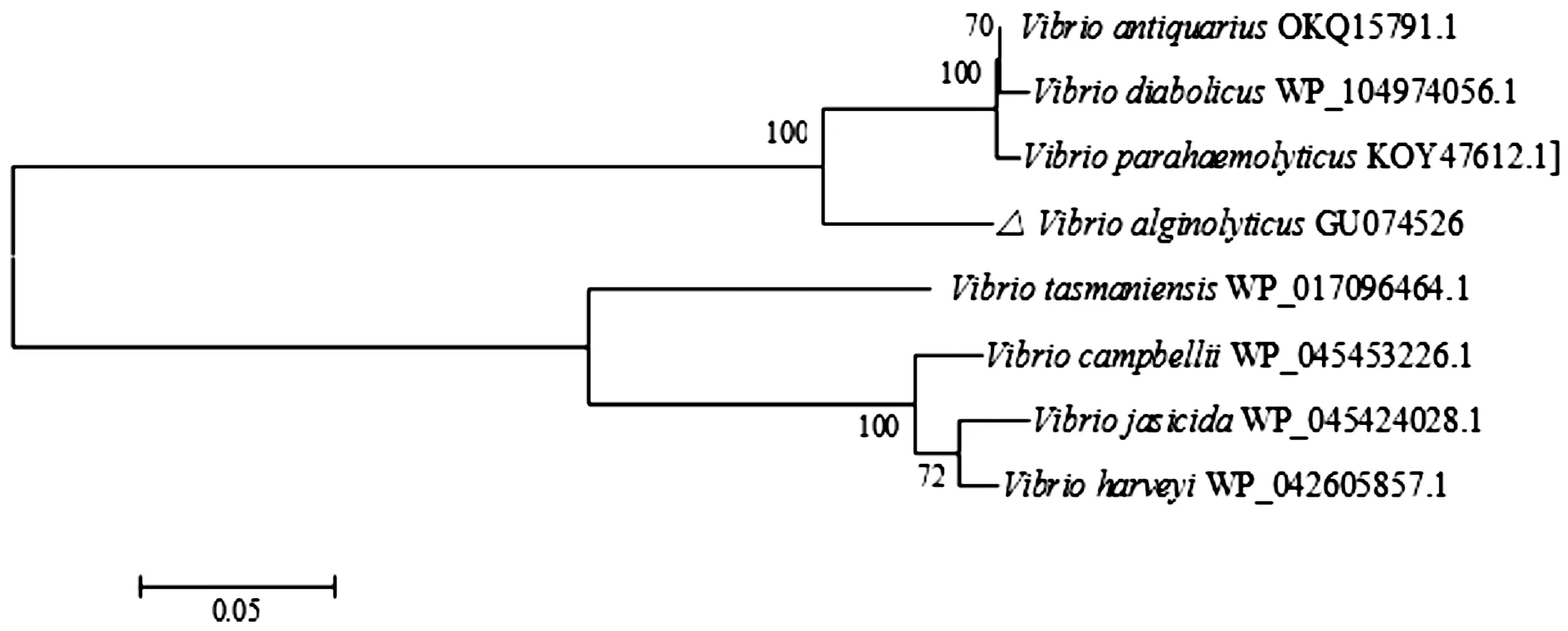
Fig.4 Phylogenetic tree of VscB amino acid sequence constructed based on NJ method
3.5 Prediction of secondary structureSOPMA was used to predict the secondary structure of VscB online, and it was found that the secondary structure of VscB protein was composed of α-helix (45.07%), β-turn (7.04%), random coil (27.46%) and extended strand (20.42%) (Fig.5).
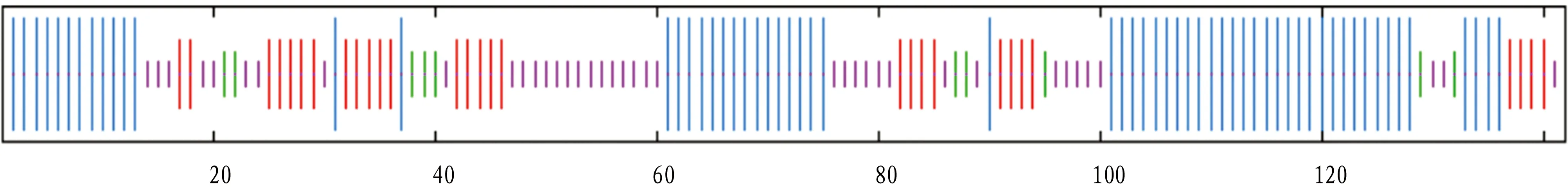
Fig.5 Prediction of secondary structure of VscB protein
3.6 Functional domain prediction for VscBThe CDD website was used to predict, and the results showed that VscB[superfamily, evalue=3.64e-51] cl08444 had a CesT family domain. CesT provided molecular chaperone function for intimal receptor (Tir) protein translocation in enteropathogenicE.coli(EPEC), giving EPEC the ability to change the morphology of host cells after bacterial attachment (Fig.6).
3.7 Tertiary structure of VscBThe amino acid sequence of VscB was modeled by Swiss-model, and the corresponding tertiary structure prediction model was obtained. The results showed that there were 3 α-helices and 5 β-turns in VscB protein (Fig.7).
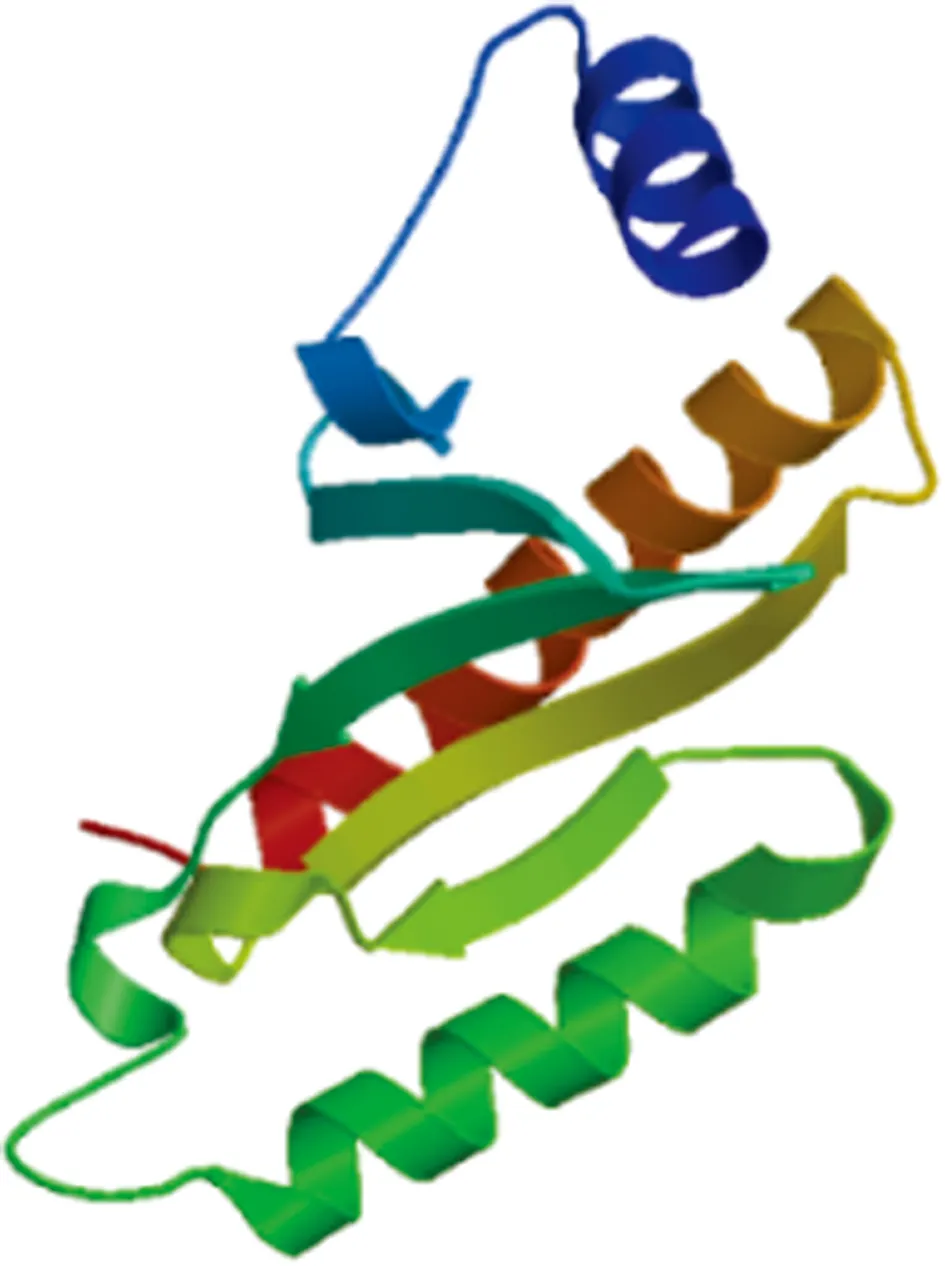
Fig.7 Prediction of tertiary structure of VscB
4 Discussion
Type III secretory system plays an important role in the process ofV.alginolyticusinfection, and VscB protein, as a component ofV.alginolyticustype III secretory system, is closely related to the pathogenesis ofV.alginolyticus. In this study, VscB protein inV.alginolyticustype III secretory system was analyzed by bioinformatics analysis. The results showed that VscB protein was not secretory protein, and had no signal peptide and no transmembrane region. It was predicted by SoftBerry-Psite that protein kinase C phosphorylation site and casein kinase II phosphorylation site existed in the amino acid sequence of the protein. Protein phosphorylation is a common regulation mode, which widely exists in organisms and can modify proteins in the body. The secondary structure of VscB was predicted online, and it was found that its secondary structure consisted of a large number of random coils, α-helices, extended strands and a small number of β-turns. Through BLAST, it was found that the protein similarity between VscB ofV.alginolyticusandVibrioParahaemolyticuswas the highest, reaching 91%. The results of multi-sequence alignment showed that VscB was relatively conservative in Vibrio. Through phylogenetic tree analysis, the corresponding proteins ofV.alginolyticusVscB,V.ParahaemolyticusandV.diabolicuswere clustered in the same subfamily, which indicated that they were close in evolutionary and biological relationship.
In recent years, there have been many reports on the effector protein of type III secretory system, while the research on chaperone of T3SS is relatively insufficient. Many effector proteins in T3SS depend on corresponding chaperones to maintain their stability in bacterial cytoplasm, and chaperones can assist effector proteins to secrete or transport[10]. Some chaperones can also be transported into host cells and participate in the intracellular functions of the host[11]. VscB belongs to Tir chaperone (CesT) family. This family consists of a large number of bacterial sequences, which are highly similar to Tir chaperones inE.coli. In Yersinia, YscB and SycN are chaperones of YopN. The secretory channel width of T3SS is only 2 nm, Yops protein needs to be maintained in a semi-folded state if it passes through the channel. It is speculated that SycN and YscB chaperones play this function and make effector protein exist stably before secretion[7].
杂志排行
Asian Agricultural Research的其它文章
- Analysis of Visualization Mapping in the Field of Brand Community Based on CiteSpace III
- Establishment of Molecular Biological Method for Identification of Bacteria by 16S rDNA and gyrB Gene
- Amplification and Bioinformatics Analysis of h-ns Gene of Vibrio alginolyticus
- Effects of Continuous Cropping and Crop Rotation on Fungal Diversity in Greenhouse Soil
- Effects of Addition of Amino Acids or Soybean Phospholipid in Diet on Slaughter Performance and Meat Quality of Pigs
- Changes in Soil Organic Matter, Nitrogen and Phosphorus Contents during Decomposition of Pear Branches
