Evaluation of Microsphere-based xMAP Test for gyrA Mutation ldentification in Mycobacterium Tuberculosis*
2023-05-10OUXiChaoZHAOBingSONGZeXuanPEIShaoJunWANGShengFenHEWenCongLIUChunFaLIUDongXinXINGRuiDaXIAHuiandZHAOYanLin
OU Xi Chao, ZHAO Bing, SONG Ze Xuan, PEI Shao Jun, WANG Sheng Fen, HE Wen Cong,LIU Chun Fa, LIU Dong Xin, XING Rui Da, XIA Hui, and ZHAO Yan Lin,#
Drug-resistant (DR) tuberculosis (TB) is a growing threat to public health[1].Fluoroquinolone (FQ)antibiotics have long been used as anti-tuberculosis drugs, and their widespread application has led to the development of resistance in clinical isolates ofMycobacterium tuberculosiscomplex (MTBC).Furthermore, TB caused by MTBC isolates that fulfills the definition of multidrug/rifampicin-resistant(MDR/RR) and is also resistant to any FQ, including levofloxacin and moxifloxacin (MOX), is referred to as pre-extensively DR-TB (Pre-XDR-TB)[2], and can become extensively drug-resistant (XDR), with additional resistance to at least bedaquiline or linezolid[2].Therefore, detecting resistance to FQ is of substantial concern for Pre-XDR-TB and XDR-TB diagnosis and treatment.
The use of conventional phenotypic culturebased drug susceptibility testing (DST) to detect DRTB fails to afford the timely diagnosis required for proper patient treatment and management.InM.tuberculosis, the principal target of FQs is DNA gyrase, a type II topoisomerase comprising two A and two B subunits encoded by thegyrAandgyrBgenes, respectively[3].FQ resistance inM.tuberculosisis primarily mediatedviagyrAmutations.Amino acids at positions 88, 90, 91, and 94 ofgyrAwere found to be the most frequently substituted in the FQ-resistant MTBC clinical isolates[4].Mutations D94G, A90V, D94A, D94N,S91P, D94Y, D94H, and G88C ingyrAwere ranked as Group 1, which means associated with FQ resistance by the World Health Organization (WHO) catalog of mutations inM.tuberculosis[5].Therefore, as an alternative to protracted culture-based methods,M.tuberculosiscan be examined for FQ resistance using alternative rapid, sensitive, and safer molecular diagnostic tools, which detect resistance by identifying mutations known to confer FQ resistance,as mentioned above.
The MagPlex microsphere-based multiple analyte profiling (xMAP) system (Luminex Corp.Austin, TX,USA) is a multiplexed detection platform capable of analyzing and reporting up to 500 different targets in a single reaction tube[6].Using labeled microspheres,xMAP offers a new platform for high-throughput DRTB diagnosis by integrating multiplex PCR amplification and microsphere hybridization technology[7].By dyeing the microspheres or beads different colors in advance and then setting the specific codes for different colored beads in the xMAP system, the different color-coded beads can be coupled with distinct oligonucleotide probes to identify corresponding DNA fragments of the PCR amplification product by hybridization.The addition of a reporter dye allows the detection of the specific hybridized PCR products in the system.In the present study, we evaluated a microsphere-based xMAP test for identifyinggyrAmutations using the xMAP platform to detect FQ resistance genotypes.A series of primers were synthesized to amplify gene fragments harboring resistance mutations in thegyrAgene, and both wild-type and mutation sites can be detected using this test (Table 1).
MTBC isolates were selected from the National Tuberculosis Reference Laboratory in China.In total,210 clinical isolates were sub-cultured on the Löwenstein-Jensen medium for analysis.Based on the broth microdilution DST, 50 isolates were MDR isolates, 39 isolates were isoniazid (INH) monoresistant isolates, 15 isolates were rifampicin (RIF)mono-resistant isolates, and 106 isolates were INHand RIF-sensitive isolates (Figure 1).All 210 MTBC isolates exhibited conclusive ofloxacin (OFL) and MOX susceptibility test results by broth microdilution: 41 (19.5%) were resistant to ofloxacin(OFL), and 43 (20.5%) were resistant to MOX.Among the 210 MTBC isolates, two main lineages were identified: 71.4% (150/210) were assigned to lineage 2 (East Asian genotype), and 28.6% (60/210) were assigned to lineage 4 (Euro-American genotype)[8](Figure 1).Notably, 42 isolates had 46 detectable mutations in thegyrAgene, as determined by wholegenome sequencing (WGS).The most prevalent drug-resistant mutations were D94G (34.8%, 16/46),followed by A90V (15.2%, 7/46), S91P (10.9%, 5/46),D94Y (8.7%, 4/46), D94N (8.7%, 4/46), D94A (8.7%,4/46), D94H (4.3%, 2/46), and D90N (4.3%, 2/46).One A288D mutation was detected in thegyrAgene,one N449K mutation was detected in thegyrBgene,and double mutations were detected in 4 (9.5%,4/42) isolates (Figure 1).Data presented in this study are available in the supplementary material(Supplementary Table S1, available in www.besjournal.com).

Table 1.Specific gyrA mutations designed for detection by the xMAP test
Compared with the broth microdilution minimal inhibitory concentration (MIC) method, the turnaround time of the xMAP test was markedly reduced, from 10–21 days to only 5 h.The sensitivity and specificity of the xMAP test for detecting resistance were 90.2% (95%CI, 76.9%–97.3%) and 98.8% (95%CI, 95.8%–99.9%) for OFL and 88.4%(95%CI, 74.9%–96.1%) and 99.4% (95%CI,96.7%–100.0%) for MOX (Table 2).

Figure 1.Maximum-likelihood tree of 210 Mycobacterium tuberculosis isolates and annotated with drugresistant information.Lineages, phenotypic drug-resistant type, and genotypic drug-resistant profile of the isolates are shown.
Of the 41 isolates exhibiting OFL-resistant phenotype, 4 were identified as OFL susceptible by the xMAP test.The single D89N mutation ingyrAwas detected in one isolate by WGS, which was listed in Group 3 (meaning uncertain significance for FQ resistance by WHO), and the MICs for OFL and MOX for this isolate were 8 and 4 μg/mL,respectively, also indicating resistance to MOX.A D89N mutation ingyrAwas identified in another isolate, which was susceptible to OFL but resistant to MOX by broth microdilution DST, and the MICs of OFL and MOX for this isolate were 2 and 2 μg/mL.As a rare mutation, its association with FQ resistance remains unclear.The MICs of the other three xMAP susceptible but phenotypically OFL-resistant isolates were 4, 4, and 8 μg/mL, respectively, and may be attributed to an unknown mechanism of FQ resistance or hetero-resistance not detected by WGS and xMAP[9].Mutations D94T and D94N in thegyrAgene were detected by WGS in two phenotypically susceptible but xMAP test resistance isolates.The MICs of these two isolates were 2 μg/mL, which is close to the breakpoint MIC and may be attributed to an unknown compensatory mutation restoring FQ susceptibility[10].
Considering 5 phenotypically MOX-resistant isolates testing MOX susceptibility using the xMAP test, 2 isolates carried no mutations according to WGS, which may be due to unknown resistance mechanisms or hetero-resistance, 2 isolates contained agyrA_D89N mutation as mentioned above, and 1 isolate contained agyrB_N499K mutation that was also listed in Group 3 by the WHO.The MIC for MOX was 1 μg/mL, indicating lowlevel MOX resistance.Considering the one isolate determined as MOX-resistant by the xMAP test but susceptible to MOX by MIC DST, genotypic resistance was attributed to a D94N mutation in thegyrAgene,as determined by WGS analysis (Table 2).The MIC for MOX was 0.25 μg/mL, which is near the breakpoint MIC for MOX and may be attributed to an unknown compensatory mutation, as mentioned above.
With WGS as the reference standard, mutations ranked in Group 1 and Group 2 were considered DR genotypically.The sensitivity and specificity of xMAP detection of resistance were 100.0 and 100.0% (95%CI, 98.9%–100.0%) for OFL and MOX, respectively.
Nevertheless, we acknowledge the limitations of this study.Firstly, the limit of detection (LOD) of FQ resistance inM.tuberculosisand the LOD for heteroresistance detection of xMAP were not tested in the present study, given that low-frequency heteroresistance is a major problem for FQ genotypic DST;this will be examined in the next step.Secondly, this was intended as a preliminary validation study of a new molecular test for detecting OFL and MOX resistance of MTBC isolates, and levofloxacin was not included for analysis because it was not included in the MYCOTB plate; the preliminary validation performance was markedly good, but more MTBC isolates with different mutations and clinical samples, such as sputum, will be utilized in future evaluation studies.
In conclusion, the xMAP test exhibited good performance in identifyinggyrAmutations inM.tuberculosis, and it could diagnose FQ resistance more accurately owing to the open-architecture platform.To expand the scope of the assay for detecting additional anti-tuberculosis drug resistance, including additional primers and probes on this open-architecture platform would enhance the use of the xMAP test in the future.
AcknowledgmentWe thank Dr.Sherry Dunbar for editing the English text of a draft of this manuscript.
&These authors contributed equally to this work.
#Correspondence should be addressed to ZHAO Yan Lin, E-mail: zhaoyl@chinacdc.cn
Biographical notes of the first authors: OU Xi Chao,male, born in 1982, MD, majoring in tuberculosis molecular epidemiology and new diagnostic tools development; ZHAO Bing, male, born in 1975, MD,majoring in tuberculosis diagnostic tools application.
Received: January 3, 2023;Accepted: February 24, 2023
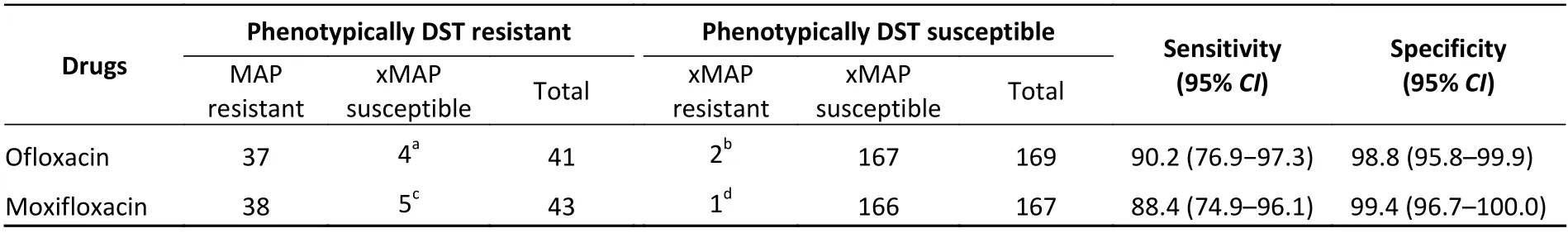
Table 2.Diagnostic performance of the xMAP test compared with that of the phenotypic DST method for detecting OFL and MOX susceptibility
杂志排行
Biomedical and Environmental Sciences的其它文章
- A Comparative Study of Blood Lead Levels in Urban Children in China: The China Nutrition and Health Survey (CNHS)2002 and 2012*
- Type 2 Diabetes and Hyperlipidemia Caused by AKT2 Gene Combined with PLlN1 Gene Mutation: A Case Report*
- Leukocyte Telomere Length and Lacunar Stroke: A Mendelian Randomization Study*
- Effect of Age and Sex on Stroke Mortality of Young and Middleaged Adults in China, 2002–2019, and Predictions to 2030
- Benefits of Mindfulness Training on the Mental Health of Women During Pregnancy and Early Motherhood: A Randomized Controlled Trial*
- rs3735664 Polymorphism Affecting ELFN1-AS1 Adsorption on miR-1231 is Associated with Colorectal Cancer Susceptibility and Tumor Stage*
