Optical Microscopy Advances Reach Sub-Nanometer Resolution
2023-03-22ChrisPalmer
Chris Palmer
Senior Technology Writer
A large team led by German scientists at the University of Göttingen has pushed optical microscopy past the milestone of single-nanometer resolution with a relatively inexpensive superresolution microscopy (SRM) technique called one-step nanoscale expansion (ONE), reporting their work in late 2022 [1].The technique employs two widely used SRM strategies, expansion and tracking fluorescence fluctuations,combining a 10-fold axial specimen expansion(1000-fold by volume)with computational analysis of fluctuations in fluorescence across up to 3000 images of a single biological sample (Fig.1).Not long after, in 2023, another German group led by researchers at the Max Planck Institute of Biochemistry in Planegg also breached the nanometer mark with a variant of standard fluorescence single-molecule localization microscopy (SMLM), describing resolution enhancement by sequential imaging (RESI), a technique employing DNA-barcoding and fluorescent tags of multiple colors [2].Although both techniques use off-the-shelf hardware and reagents, the equipment needed for RESI is much more expensive.
‘‘This is extraordinary.Going down to nanometer or even angstrom optical resolution had previously been deemed impossible,”said Robert Prevedel, a microscopy expert and group leader at the European Molecular Biology Laboratory in Heidelberg, Germany.‘‘Just ten years after the Nobel Prize for super-resolution microscopy achieving around 50-nanometer resolution, we are down to a nanometer or less.”
That 2014 Nobel Prize in Chemistry was awarded to three principal innovators in SRM [3]—Eric Betzig(Janelia Research Campus,Howard Hughes Medical Institute, Ashburn, VA, USA), Stefan W.Hell (Max Planck Institute for Biophysical Chemistry, Göttingen,and German Cancer Research Center, Heidelberg, Germany), and William E.Moerner (Stanford University, Stanford, CA, USA)—‘‘···for circumventing a basic law of physics and enabling microscopes to peer at the tiniest structures within living cells.”
Just 25 years ago, life scientists peering down the barrels of optical microscopes could clearly make out objects right at the diffraction limit, a fundamental law of optics stating that image resolution cannot exceed half the wavelength of the light being viewed.For visible light,that limit is about 200 nm,meaning many subcellular structures were too small to observe in meaningful detail.Various forms of electron microscopy (EM), first invented in the 1930s, have far surpassed this limitation, revealing cellular structures and molecular details at the sub-nanometer level.But while EM generates images from sample-damaging beams of electrons instead of particles of light to achieve high resolution,its need for thinly sliced, specially prepared specimens placed in a vacuum precludes imaging of live samples.Today, thanks to the creative approaches pioneered by Betzig, Hell, Moerner, and others, engineers have built an increasingly sophisticated array of super-resolution optical microscopes far less expensive than their EM counterparts, some even smart enough to use artificial intelligence(AI)to autonomously collect data around the clock and analyze it in real-time.
A small number of SRM techniques first developed in the late 1990s achieved 20 nm resolution using combinations of specially engineered excitation light sources,fluorescent dyes, highly sensitive detectors, and reconstruction algorithms [4].SMLM, one such method, involves labeling all the copies of a molecule of interest with a fluorescent tag and then causing these tags to light up,but just a few at a time to prevent tags of adjacent molecules blurring together.Over many repetitions of this process, the positions of molecules of interest can be determined,and their arrangement reconstructed by combining the positions of each into a single data set (Fig.2) [5].However, while widely used, the new techniques were relatively expensive, requiring specialized sample preparation and a confocal microscope, which can cost from 100 000 to 250 000 USD.
The year 2015 saw the introduction of expansion microscopy,a new method for achieving SRM on the cheap [6].With expansion microscopy, a fluorescently labeled sample is crosslinked to a hydrogel, which serves as a scaffold for the subsequent process.The hydrogel, molecularly like the absorbent material in a disposable diaper,expands isotropically when exposed to water,causing the sample to enlarge in all three dimensions.The expansion physically enlarges the sample, circumventing the diffraction limit by effectively separating the fluorescent labels to allow higher-resolution imaging.When used with a conventional fluorescence microscope costing from 10 000 to 20 000 USD,standard expansion SRM can resolve structures initially as small as 70 nm.
Because it is relatively inexpensive and easy to perform,expansion microscopy has spread widely,with more than 1000 laboratories publishing research based on its use [5].In fact, an entire cottage industry has developed around expansion microscopy,with dozens of variants released in the past several years,including a version that allows the expansion factor to be thermally adjusted in a reversible manner[7],a high-throughput version for assessing 96 samples at once [8], and another yielding an 8000-fold volume expansion that allows ultrastructural features to be visualized with simple magnification optics alone [9].And while expansion microscopy could initially only visualize proteins, new variations can track lipids, glycans, proteins, DNA, RNA, and small molecules[10], as well as characterize chromatin and transcription factors in vivo to study nuclear organization at the nanoscale [11].
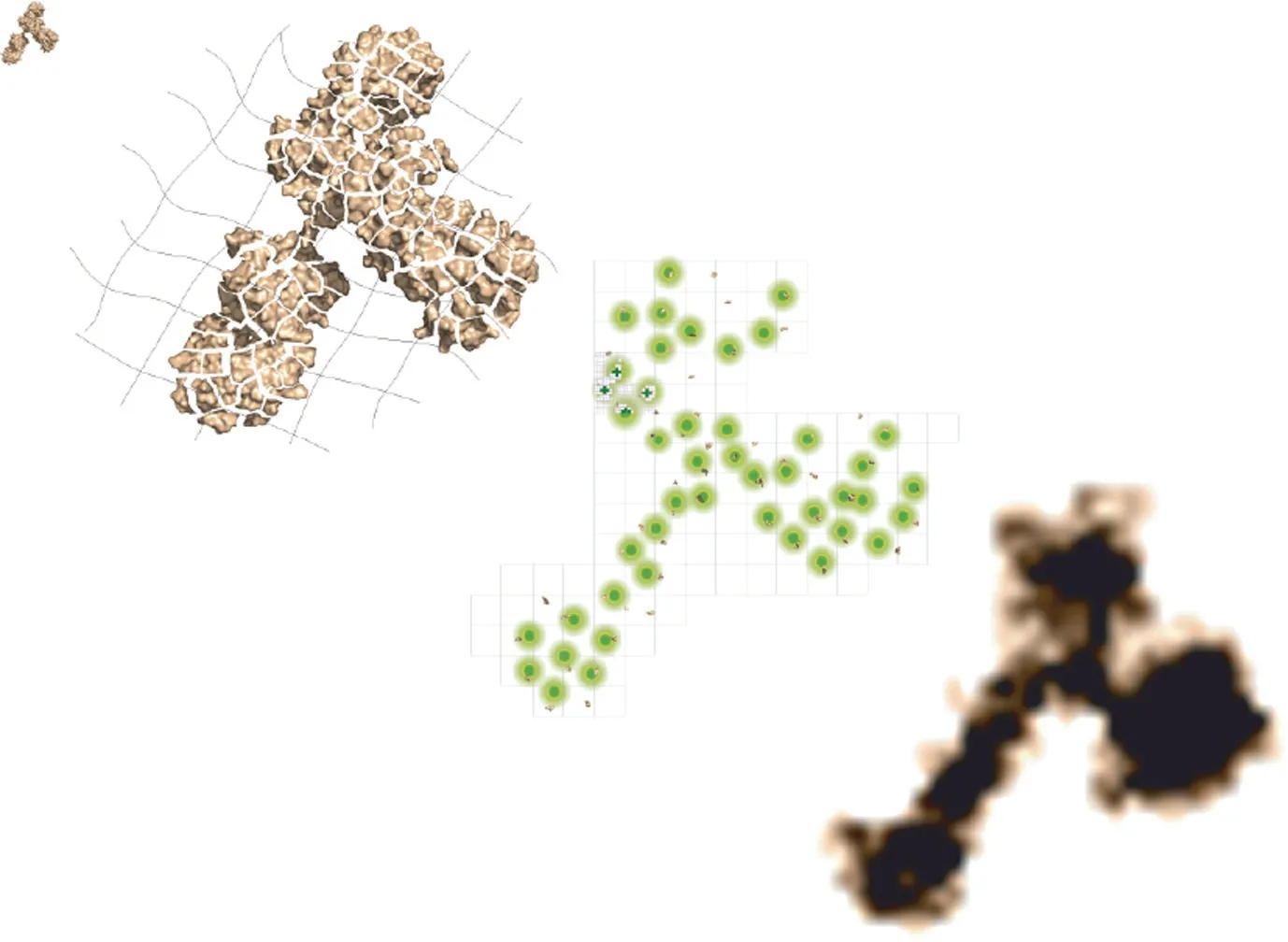
Fig.1.A general schematic of the ONE microscopy approach.Biological samples are linked to a gel that undergoes 10-fold axial expansion upon exposure to water.Using any desired imaging system to detect fluorescence signal fluctuations, up to 3000 images are combined using a computational algorithm (ONE plugin) to assemble a final super-resolved image.Credit: Ali Shaib, with permission.
These advances set the stage for ONE, which the researchers used to capture images of γ-aminobutyric acid type A (GABAA)receptor that closely resemble much-higher-resolution cryogenic(cryo)-EM and X-ray crystallography maps of the protein (Fig.3)[1].However, the method cannot match the resolution of cryo-EM, which can reveal near-atomic-level details smaller than 0.2 nm in some cases, though at a significantly higher cost and not with every type of protein structure.In contrast, ONE offers a straight-forward, simple method for obtaining insights into the structure of just about any molecule; it is also compatible to use with now-antiquated 1990s-era fluorescence microscopes.
‘‘Cryo-EM is a powerful technique that comes with a high cost.One needs a lot of resources,skills,maintenance,and time to run it and analyze the results.Even if you have a well-funded lab, you cannot use cryo-EM for every question you have,” said Ali Shaib,first author of the report describing ONE and a group leader at the Institute of Neurophysiology,Department of Prof.Silvio Rizzoli,University Medical Center Göttingen, in Germany.‘‘So, why not compliment your work with an intermediate step using ONE microscopy?”
While inexpensive options for SRM like ONE could help more scientists perform cutting-edge research, so may a suite of ‘‘smart microscopy” tools intended to relieve investigators of the tedium associated with the time-consuming data acquisition and analysis involved in high-resolution imaging of biological processes.Despite substantial advances in microscope optics and computational resources,researchers still conduct many microscopy experiments much as they did decades ago,zeroing in on samples one by one,waiting patiently for an event of interest to occur,and recording just enough repetitions to achieve scientific rigor.‘‘Microscopes are,for the most part,passive machines,”Prevedel said.‘‘But in the future, there will be a big drive to make microscopy smart and autonomous.”
Prevedel heads one of many research groups developing custom-built, smart microscopes that rely on delicate, highly responsive motors and optics coupled with machine-learning approaches to peer deeper into tissues than ever before and to zoom in at just the right time to capture fleeting moments in the life of a cell.‘‘Cell biologists and developmental biologists like to observe cells or even entire organisms as they develop in time.Very often, there are minutes-to-hours where nothing happens,and when it does,it is only in one corner of the specimen.So,they are hunting for rare events in time and space,” Prevedel said.‘‘Smart microscopes can adapt their acquisition rate, field of view,resolution, and whatnot to capture the most information possible without the operator sitting there steering the microscope for hours at a stretch,which helps when biologists have to obtain hundreds, maybe even thousands, of replicate images for a study.”
Using automated tools to save images to disk only when phenomena of interest occur also requires far less disk space than letting the microscope run at its highest frame rate for up to days at a time,drastically slashing the time needed for subsequent analysis.‘‘In addition to the time savings, [autonomous microscopy] offers more objective acquisition of biological samples, reducing biases that human microscope operators may have,”said Anne Carpenter,a computational biologist and senior director of the Imaging Platform at the Broad Institute of Massachusetts Institute of Technology and Harvard University in Cambridge, MA, USA.Carpenter developed CellProfiler, open-source software for measuring observable traits in cell images that is based on machine-learning algorithms commonly used in facial recognition.Since its release in 2005,the software has been cited in more than 16 000 publications[12].‘‘In the extreme,[the software]might uncover findings a biologist was not looking for and might have missed,” Carpenter said.

Fig.2.Left:A conventional diffraction-limited image of DNA in the nucleus of a HeLa cell labelled with a fluorescent DNA-binding dye.Right:Super-resolution(50 nm)image of the same HeLa nucleus captured with an SRM method called fluctuation-assisted binding-activated localization microscopy(fBALM).The middle panels show the indicated sections of the larger panels at 500 nm resolution.Credit: Andy Nestl (CC BY-SA 4.0).
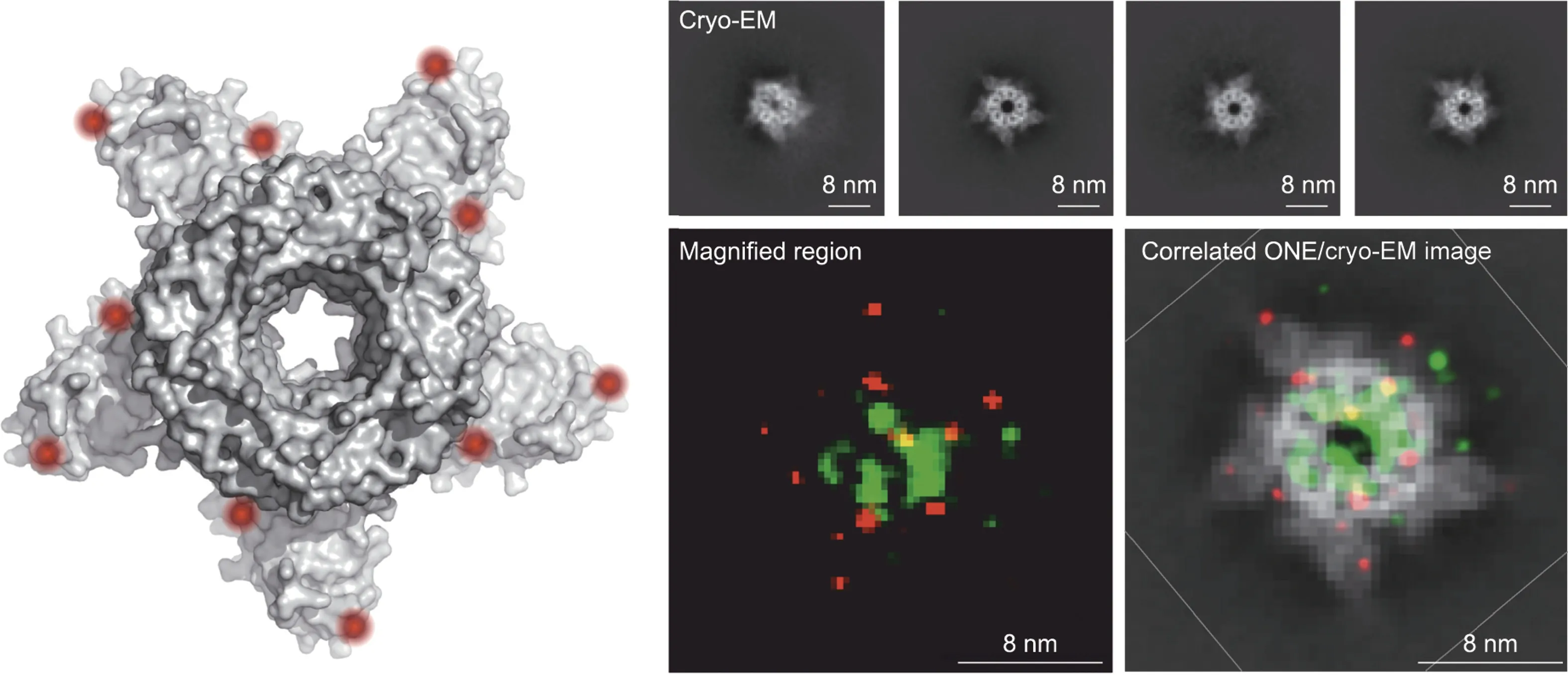
Fig.3.Left:A cryo-EM reconstruction of GABAA,a neurotransmitter receptor found throughout the brains of many animals.Right top:Individual cryo-EM images of GABAA captured at different angles.Right bottom: ONE microscopy image of GABAA (left) and the same image correlated with cryo-EM image (right).Credit: Ali Shaib, with permission.
Over the past several years, Prevedel and his colleagues have engineered a microscope combining three-photon fluorescence imaging, which is used to probe inside tissues, with an adaptive optical system, a technology first developed for astronomy.The system works by directing light using mirrors and lenses made of deformable membranes instead of rigid materials.Software drives real-time changes to the membranes’ shapes in response to variations in the sample,bending light in specific ways to peek through surface structures and continuously capture high-quality images.By also timing image acquisition to an animal’s heartbeat,Prevedel’s system can visualize and record the activity in cells nearly 1.5 mm beneath the brain’s surface in a region called the hippocampus.That is 0.5 mm deeper than previous efforts [13].
Autonomous microscopy can also help minimize the damage that biological tissue experiences during imaging.Ilaria Testa, an associate professor of applied physics at the Royal Institute of Technology in Stockholm, Sweden, and her team combined two microscopy approaches: a fluorescent wide-field microscope and a form of SMR known as stimulated emission depletion(STED)that requires intense illumination.When the team’s software detects a change in fluorescence from the wide-field view, the system switches automatically to the higher-resolution STED mode,thereby limiting the sample’s exposure to damaging bright light.This hybrid tool allowed Testa and colleagues to observe—with nanometer precision—how nerve cells reorganize their synaptic vesicles after releasing calcium [14].
While researchers like Prevedel and Testa have developed their own smart microscopy systems,a few out-of-the-box solutions are available to labs not wanting to build from scratch.For example,the AutoPilot framework developed by scientists at Howard Hughes Medical Institute’s Janelia Research Campus in Ashburn,VA, USA, enables time-lapse, intracellular imaging of the development of zebrafish, fruit fly,and mouse embryos [15,16].The hardware and software system,which takes a couple of weeks to set up,manages position and three-dimensional orientation of the sample as well as acquisition parameters to optimize speed, quality, and imaging consistency over time [17].
Other off-the-shelf hardware and software systems for automating microscopy have been developed and are offered by powerhouse imaging equipment vendors Zeiss (Oberkochen,Germany),Thermo Fisher (Waltham,MA, USA), and Nikon (Tokyo,Japan) [18].Startup companies are also making autonomous microscopy available to more labs, including Celly.AI (Covina, CA,USA), which allows a conventional optical microscope to operate autonomously via a motorized stage and AI software running on a cell phone attached to the eyepiece [19].
‘‘One of the biggest challenges is being able to control multiple necessary automated components of the microscope—they all have different software and firmware that are being constantly updated,” said Carpenter, citing the open-source MicroManager app software which manages the task of maintaining drivers for many common components so that biologists need only focus on putting together automated workflows[20].‘‘That project has done a great service for the scientific community,” she said.
From major advances in optics and AI control to clever combinations of existing techniques to tools for helping components work together optimally, researchers will continue to benefit as these technologies are incorporated into commercial microscopes.‘‘It will be exciting to see individual automated features gradually added over time,” Carpenter said.‘‘I suspect it will not be one major transition, but instead tasks will get incrementally easier as biologists take advantage of convenient, built-in automated tools.”
杂志排行
Engineering的其它文章
- Global Top Ten Engineering Achivements 2023
- 2023 Global Engineering Fronts
- Will Massive Appetite for Minerals Stall Clean Energy Transition?
- International Correlation Research Program: Cross-Fault Measurement for Earthquake Prediction
- A Systematic Perspective on Communication Innovations Toward 6G
- Data-Driven Modeling of Maritime Transportation: Key Issues,Challenges, and Solutions
