Identification and Expression of the Conotoxin Homologous Genes in the Giant Triton Snail (Charonia tritonis)
2023-03-17JIAHuixiaZHANGGegeZHANGChenglongZHANGHuaYAOGaoyouHEMaoxianandLIUWenguang
JIA Huixia, ZHANG Gege, ZHANG Chenglong, ZHANG Hua, YAO Gaoyou,HE Maoxian, and LIU Wenguang, *
Identification and Expression of the Conotoxin Homologous Genes in the Giant Triton Snail ()
JIA Huixia1), 2), ZHANG Gege1), 2), ZHANG Chenglong4), ZHANG Hua1), 3), YAO Gaoyou1), 2),HE Maoxian1), 3), and LIU Wenguang1), 3), *
1),,,,510301,2),100049,3),511458,4) Sansha Marine and Fisheries Bureau, Sansha 573199, China
Conotoxin (CTX) is a small peptide toxin, and plays crucial role in anesthetizes prey, preys and defends competitors.is the natural predator of the crown of thorns starfish (CoTS) and the protector of coral reefs. It plays an important role in the coral reef ecosystem. However, the types of toxins produced by tritons and the molecular mechanisms of toxin secretion ofare unknown. In the present study, the four conotoxin homologous genes (s) fromwere identified. Species and conotoxin superfamily phylogenetic tree indicated that(CL2216.Contig2) and(Unigene 58438_All)belong to the C superfamily,(Unigene 66414_All)belong to the V superfamily, and(Unigene 21408_All)belong to the B1 super- family.swere highly expressed in salivary gland, liver and digestive gland. The predation control experiment revealed that the expressions ofs were significantly different in salivary gland, liver and digestive gland. The results indicated thats may par- ticipate in the process ofpredating CoTs, and provided a scientific basis for further studying the toxins secretion mecha- nism of.
; toxin; tissue distribution; predation experiment
1 Introduction
The Pacific triton, also known as the giant triton snail, is one of the largest carnivorous gastro- pods in coral reef ecosystem(Zhang., 2013). It is wide- spread in tropical waters of the South China Sea(Zhang., 2021). It is also the primary predator of the crown of thorns starfish (CoTS), which is the main cause of damage to many coral reefs(Poulsen, 1995; Kang and Kim, 2004; Cowan., 2017). With the aims of protecting the coral reef ecosystem and biodiversity us- ing eco-friendly products, it is important to collect as much information as possible on the biology of, such as the predation mechanism.
Previous research has shown that many gastropods secrete acidic venom or toxin to paralyze their prey. For ex- ample, the, which is also known as at- lantic triton snail, utilizesits toxins to immobilize prey, and the toxins are secreted from foot or mouth. The Atlantic conch (), secretes toxins from itsfeet or mouth to immobilize prey(Bandel, 1984). Thecan cause the starfishparalysis instantaneously, becauseits proboscis can secrete toxins (Endean, 1972).produces hundreds of different short peptides termed conotoxin to access their prey including capture and digestion of prey, and defendagainst prey(Kaas., 2010). In, its proboscis stretches out to contact the prey, then penetrates the prey toinject toxins or acidic saliva, and quickly paralyzes the prey(Chesher, 1969; Percharde, 1972; Bose., 2017b). It has been reported that the acidic material can promote pene- tration of the calcareous body wall of the preyor trauma- tize their prey to avoid predation(Garcia Ortega., 2011). However, there are few reports on whether the acidic ma- terial is atoxin. In addition, no studies have re- ported toxin classification, identification, diversity and co- notoxin homologous genes of
In the present study, three-dimensional structure ofswere constructed and the disulfide bridge were predicted by using the amino acid sequence of conotoxin as a model. Thephylogenetic treeshowedthats Clus-tered with the C, V, B1 conotoxin superfamily.To investi-gate the possible biological roles ofs, the predation control and tissue distribution were detected, the results re- veal thes may take part in the predation process ofand will provide further insight and a theoretical ba- sis for revealing the conotoxin action and secretion me-chanism of.
2 Materials and Methods
2.1 Sample Collection
(=9) were collected from the South China Sea, which were kept active and healthy in a cement pool with seawater. Only three were in a resting stage and theirmantle, foot muscle, proboscides, tentacles and salivary gland tissues were extracted. The mantle, foot muscle, pro-boscides, tentacles, salivary gland, digestive gland, gonad and liver tissues were from the other sixin cap- ture control and treatmentgroups. All tissues were stored at −80℃ after quickly frozen in liquid nitrogen.
2.2 Nucleotide Sequence and Bioinformatic Analyses
Nucleotide sequence similarities were examined by BLAST software (http://smart.embl-heidelberg.de/). Using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) to analyze the fourgenes open reading frame (ORF).TheSMART online tool (http://smart.embl-heidelberg.de/) wasused to analyzed the amino acid sequence and structuraldomain ofs. The SWISS-MODEL online tool (https://www.swissmodel.expasy.org/interactive) and PyMOL wereused for predicting and polishling the three-dimensionalstructure ofs amino acids. Multiple sequence align-ment ofs amino acid sequences was detected using ClustalX and CLC Main Workbench 6.0 software to high- light regions of conservation. With Molecular EvolutionaryGenetics Analysis (MEGA) 6.0 software (Tamura, 2013)and Neighbor joining method of 1000 bootstrap cycles, a phylogenetic tree was constructed.
2.3 RNA Extraction and Reverse Transcription from All Tissues
To analyze the expression ofs in different tissues,mantle, foot muscle, proboscides, tenta- cles, salivary gland, digestive gland and liver tissues were collected. RNA was extracted from all tissues by using theuniversal RNA extraction kit (Magen). Then, using the re-verse transcription kit (Toyobo) with gDNA Remover to perform cDNA first-strand synthesis.
individuals in the predation control experiment were divided into two groups,., control group withoutpredation and treatment group during predation. In the ce- ment pool feeding CoTS, threethat had not eaten CoTS were captured and the remainingwere in- cluded in the post feeding group during predation on CoTS. The mantle, foot muscle, proboscides, tentacle, salivarygland, digestive gland, gonad and liver tissues were collect- ed from the different groups for quantitative real-time PCR (qRT-PCR) assay.
2.4 qRT-PCR Analyses of CTXs Expression
qRT-PCR was employed to assay the distribution ofsin different tissues and analyze thes expression dif-ference in the predation control and treatment groups. Fourpairs of primers (-realtime-F/R;-realtime-F/R;-realtime-F/R;-realtime-F/R) were designed based on the specific sequence of the fourgenes by primer premier 5software.
Each sample were tested in a 384 well plate and the am- plifications were performed in a total volume of 10μL, con- taining 5μL SYBR Green Realtime qRT-PCR Master Mix (Toyobo), 0.3μLforward primers and 0.3μL reverse pri- mers in distilled water (10μmolL−1), 3.4μL ofdistilled wa- ter and then add 1μL template cDNA that was diluted in double distilled water. The reaction program was set 94℃predenaturation for 3min, 98℃ denaturation for 10s, the annealing temperature was set 55℃/59℃/60℃ for 25s, pre-denaturation was cycled one time, and 45 denaturation an- nealing extension cycles.gene was employed as the internal reference gene.All tissues were set three bio- logical and technical replicates. Take the 2−ΔΔCtas the cal- culation method. Statistical analysis was performed withT- test by Prism 8.0. Differences were considered significant when0.05. Allsequences of the primers were listed in Table 1.
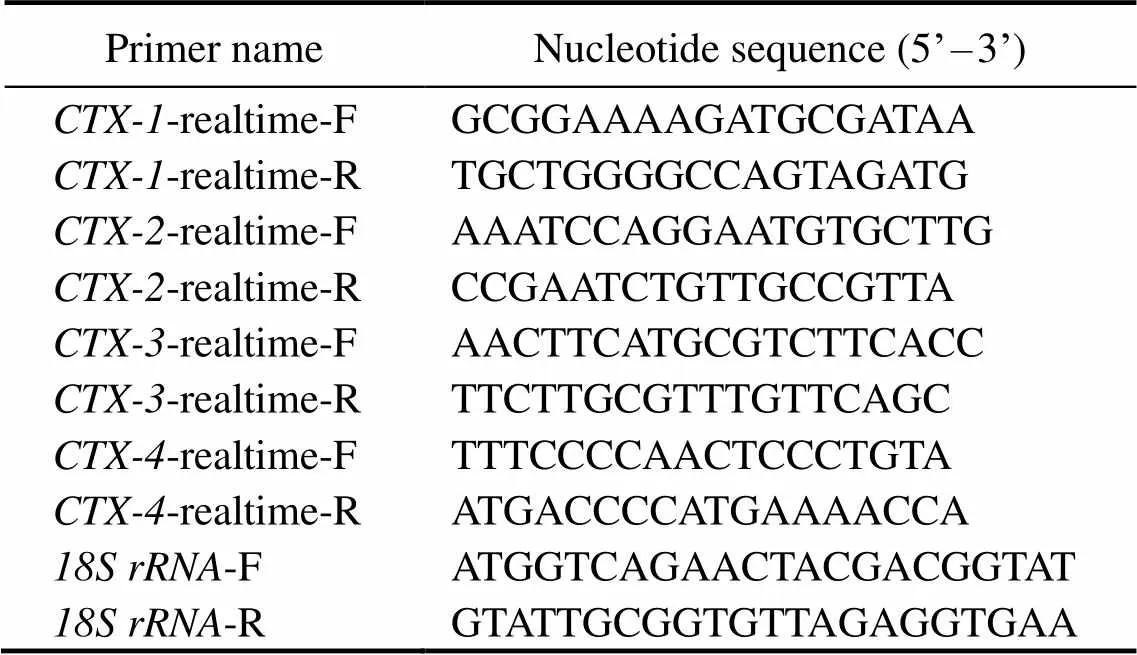
Table 1 Universal and specific primers
3 Results
3.1 CTXs Identification
Thessequences were selected from transcriptome data of the article (Zhang., 2021) about.s were identified as conotoxin homologous genes refer to the four aspects Fig.1(Kaas., 2010) (Sections 1, 2, 3, 4). Evaluate the superfamily classification based on the similarity between the CTXs protein signal peptide sequence and the reported conotoxin signal peptide sequence (Fig.1, Section 2). Next, the mature sequences formation the diffe- rent disulfide bonds (Fig.1, Section 3). Then CTXs amino acids fold to form a three-dimensional structure (Fig.1, Sec-tion 4 and Fig.2).
3.2 Sequence, Structure and Phylogenetic Tree Analysis
The protein-protein BLAST usedto match species with the highest similarity toCTXs, sequence anno- tations and amino acid sequences based on putative con- served domains and e-value. Analysis of CTX-1 revealed 43.55% identity with(accession no. AXL_95724.1), CTX-2 revealed 54.84% identity with(accession no. AMP_44780.1), CTX-3 revealed 74.50%identity with(accession no. ALM_87513.1), and CTX-4 revealed 80.00% identity with
(accession no. BAS_25475.1). The four CTXs have diffe- rentcysteine residues and disulfide bonding mode. CTX-2 has the same domain with Kunitz domain inhibitor show- ed the disulfide bonding pattern of Cys5(C1)-Cys55(C6), Cys14(C2)-Cys38(C4), Cys30(C3)-Cys51(C5). CTX-3 re-vealed the disulfide bonding pattern of Cys1(C1)-Cys57(C3),Cys48(C2)-Cys114(C6). CTX-4 has three cysteine residues suggested the disulfide bonding pattern of Cys9(C1)- Cys31(C3) and CTX-1 has five cysteine residues without disulfide bonding pattern (Fig.2). The three-dimensional structure of CTXs also conform to this result (Fig.2).
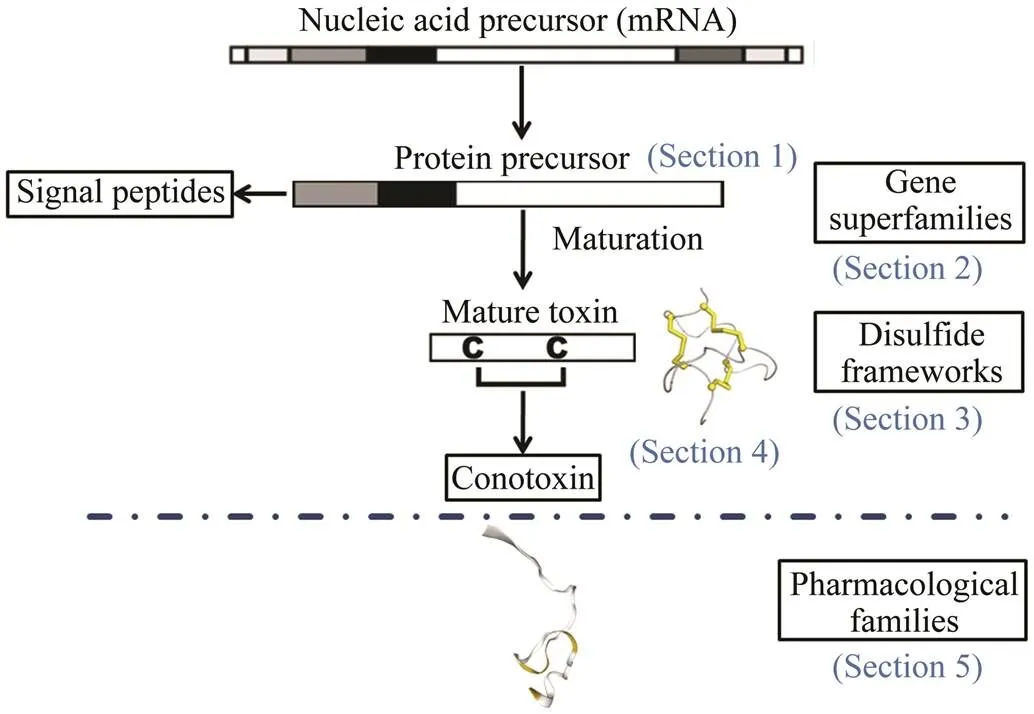
Fig.1 Schematic diagram of CTXs identification and clas- sification process. The process of evaluating the super- family classification based on the similarity between the CTXs protein signal peptide sequence and the reported conotoxin signal peptide sequence (Section 2).The cys- teine arrangement of CTXs were predicted by Conoser- ver (Section 3).The three-dimensional shape of disulfide bonds after folding (Section 4). The pharmacological fa- milies were defined based on the targeted receptor and the effect in Section 5.
Sequence alignment of CTXs showed poor sequence con- servation and different cysteine residue positions (Fig.3a). The phylogenetic tree showed that the sequence identified in(accession no. XP_041354957.1) was orthologous to the identified CTX-2 in. These- quence identified in(accession no. AXL 95723.1) was orthologous to the identified CTX-1. These- quence identified in(accession no. ALM 87513.1) was orthologous to the identified CTX-3. These- quence identified in(accession no. BAS 25475.1) and CTX-4 were clustered into one branch (Fig.3b). The tree shows that the sequence identification of CTXs and vertebrates were relatively low, but were closely re-lated to gastropod mollusks. This phylogenetic tree ana-lysis indicated that CTXs were conserved during evolu-tion.
Fig.2 3D structure and disulfide bonding pattern ofCTX-1, CTX-2, CTX-3, CTX-4.
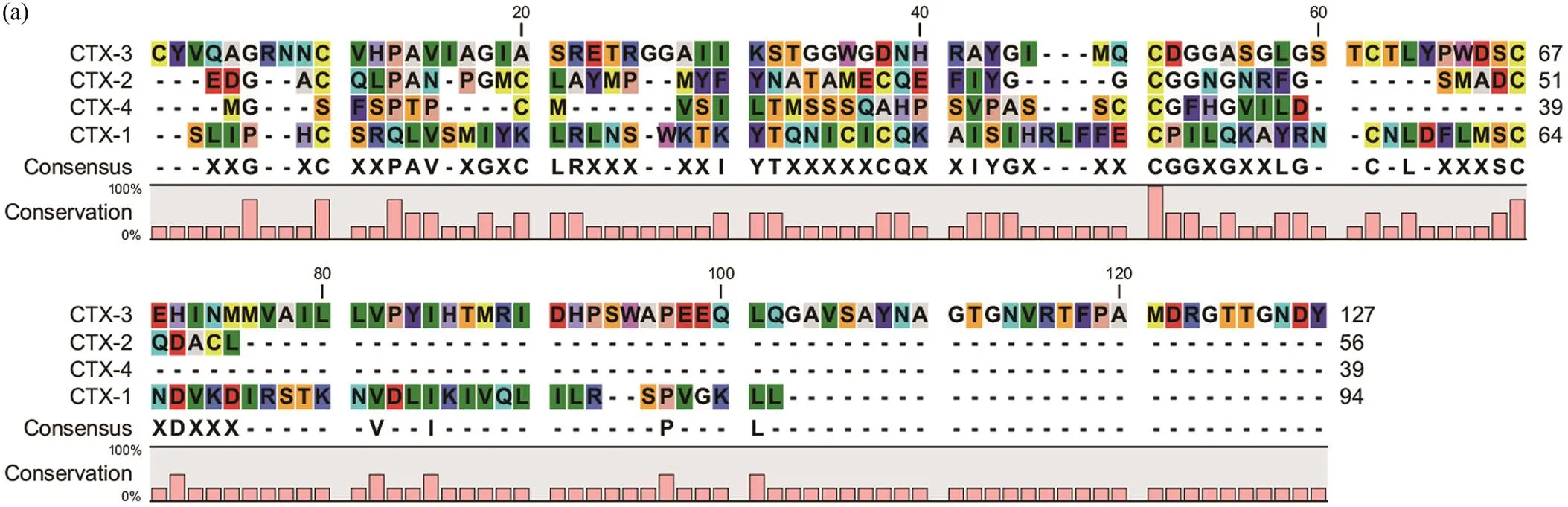
Fig.3 (a)Amino acid sequence alignment result of CTXs. The height and letter size of the pink bar graph were positively correlated with the degree of conservation of the corresponding amino acids.

Signal peptide sequenceof CTXsin Table 2 could be as- signed to the gene superfamily present in the ConoServer database(Biggs., 2010; Luo., 2013).The phylo- genetic tree of CTXs and superfamilies (D, L, C, A, S, E, M, V, B1, I2) were analyzed. This tree demonstrated that the CTX-1 and CTX-4 sequences ofand the C superfamily were clustered into one branch. CTX-2 sequenceand the V superfamily were clustered into one branch. CTX-3 sequence and the B1 superfamily were clustered into one branch.The phylogenetic treeof superfamily supported theConoServer prediction results (Fig.3c).
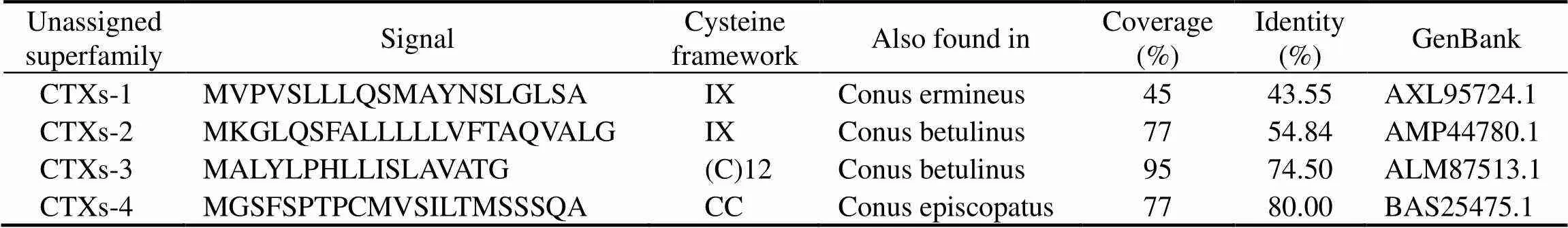
Table 2 Results of CTXs cysteine framework and BLAST
Note: The cysteine framework prediction was constructed using ConoServer.
3.3 Expression of CTXs in Tissues and Predation Controls
The tissues distribution resultsshowed thats were highly expressedin liver, salivary gland and digestive gland(Figs.4a, 4b, 4c, 4d).,andwere high-ly expressed in liver (Figs.4a, 4c, 4d).expression in salivary gland was two times higher than that in liver (Fig.4b).was highly expressed in digestive gland and liver, but low in salivary gland (Fig.4c). Moreover,ex- pression in salivary gland and liver was three times higher than in digestive gland (Fig.4d).
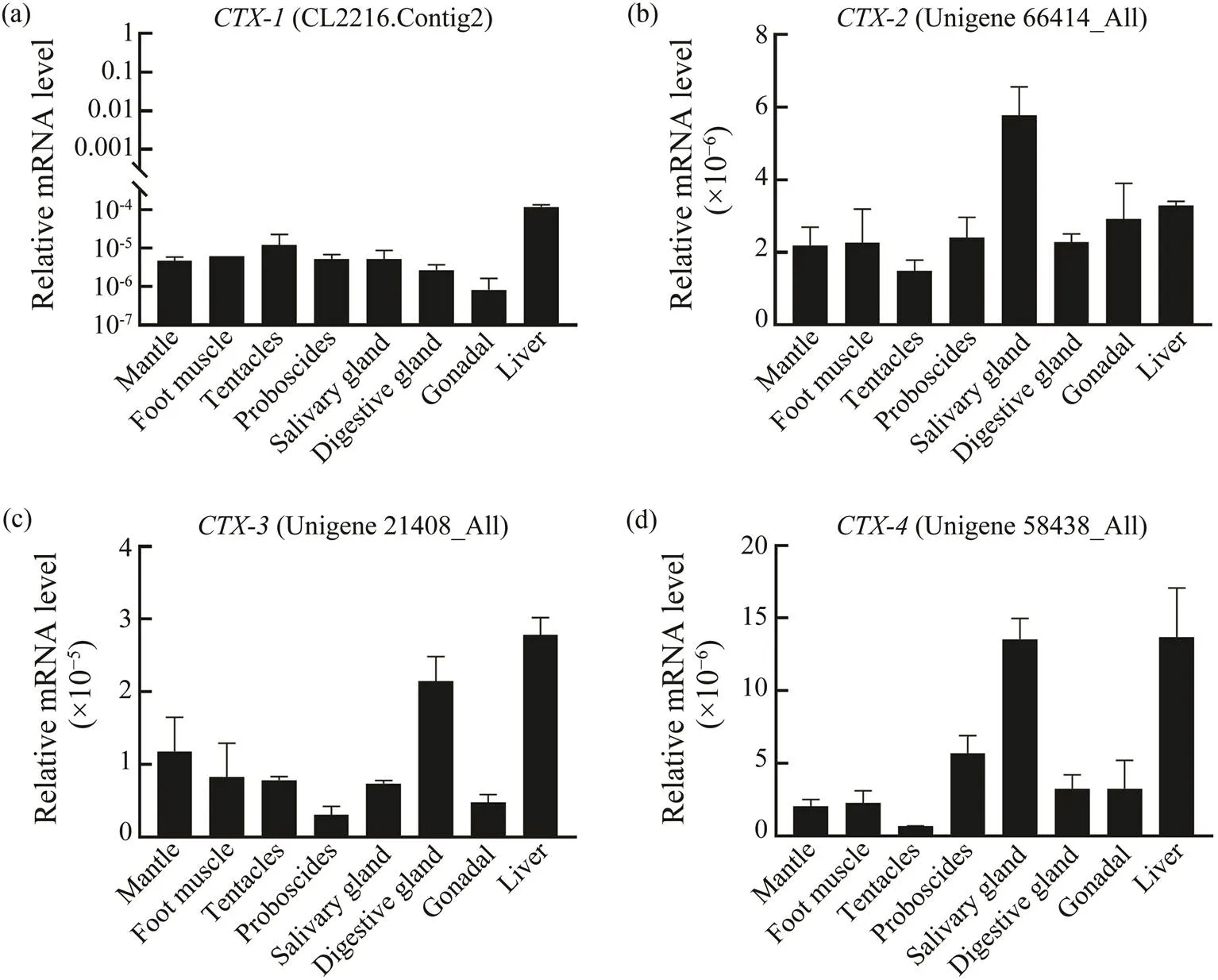
Fig.4sexpression levels in different tissues of.(a) Expression analysis of;(b) Expression analysis of; (c) Expression analysis of; (d) Expression analysis of
To analyze the expression ofs in response to preda- tion control in the salivary gland, digestive gland and liver, qRT-PCR was performed. In the salivary gland, digestive gland and liver, expression ofwas down-regulated in the control group, but in the treatment group,ex-pression was up-regulated significantly at salivary gland (0.02) (Fig.5). Increased expression oftreat- ment groupsuggested thatmight have different re- sponse mechanisms when involved in paralysis and inges- tion CoTS. However, expressions of(0.02),(0.05) and(0.02) were significantly up-re- gulated in liver but down-regulated in salivary gland com- pared to the control group (Fig.6). Expression of(0.02) in digestive gland was also significantly up-regu- lated and,andwere also up-regulated with no significant difference(Fig.7).
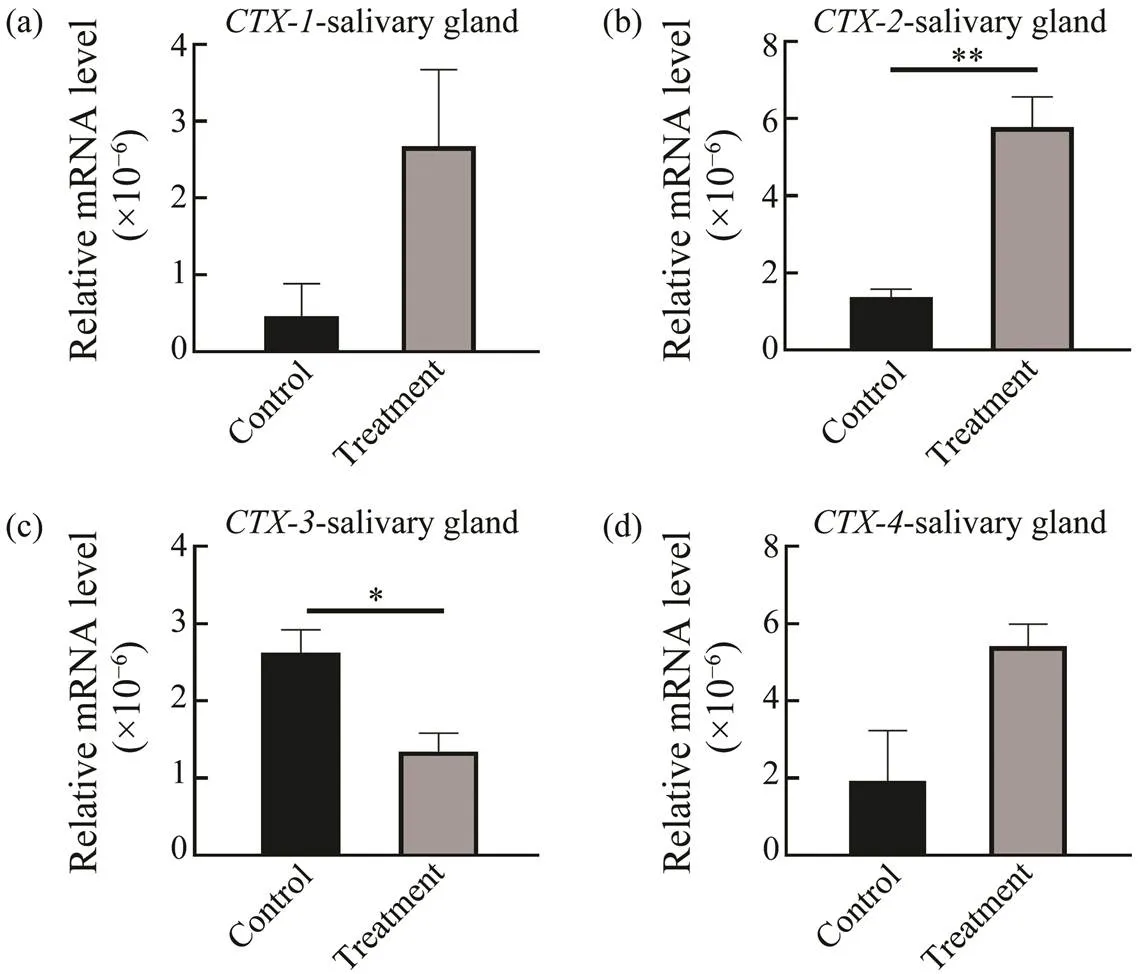
Fig.5 CTXsexpressions in salivary gland at predation control and treatment groups. *P<0.05, ** P<0.02.

Fig.6CTXs expressions in liver at predation control and treatment groups. *P<0.05, ** P<0.02.
4 Discussion
It has been reported thatcan secrete venom that can paralyze the prey, such as fish, insects, and snails (Luo., 2007; Safavi-Hemami., 2011). However, the sa- livary gland ofcan produce venom, which help predation, immobilize prey and neutralize the intrinsic to- xin of CoTS (Bose., 2017a, 2017b). Previous studies have showed that the venom secreted byandplays the crucial role in paralyze prey, and ω-MVIIA derived from the venom ofwas confirmed to treat chronic pain in cancer(Miljanich, 2004; Lynch.,2006; McGivern, 2007; Bose., 2017b).is thenatural enemy ofand its predation mechanism has been the focus of our attention. Taking into account the role of CTXs in,,and,we speculate thats may play a role in different predation periods of
Studies revealed that the composition and structure ofconotoxin vary greatly among species, including,and(Dutertre., 2013; Peng., 2016; Phuong., 2016; Li., 2017; Ro- binson., 2017). In the present study,s were iden- tified to have different types of cysteine framework based on conserved sequence (Abalde., 2018), Because the prediction by ConoServer is based on mature peptides se- quence rather than specific conserved sequence (Kaas., 2008, 2010, 2012; Brown., 2016), the CTXs have dif- ferent cysteine arrangement and are inconsistent with pre- diction. Presently there are nearly no referable reports on the pharmacological functions ofs inHope- fully our next research will focus on such function.
The phylogenetic analysis results showed that CTXs ofwere conserved inevolutionary relationship. The mantle, foot muscle, proboscides and tentacle tissueswere confirmed to participate in thehunting behavior of(HallandKingsford, 2016;Bose., 2017b; Hall., 2017).Especially, the expression difference of,,antle, foot muscle, proboscides, tentacle,salivary glands, digestive glands and liver tissuesofhas been strongly proved,which means thats mighthave different functions and response mechanisms when participating in predation.
The predation control experiment showed that signifi- cantly mores were secreted during predation com-pared with the predation control group, which showed thats possibly play a role in predation.The salivary gland, liver and digestive gland as the important digestive systemexist in almost all gastropods mollusks(Cui., 2000),which can synthesize and secrete digestive enzymes.was highly expressed in salivary gland in the control group, which was consistent with the result that the salivary gland ofcan secrete venom. However,also ex- pressed in liver and digestive gland compared with the con- trol group, which means that thes ofmight play an important role in preying on CoTS. The expression levels ofs were inconformity with the control group,which may be due to the different functions ofs in tissues. The expression difference in predation control re- vealed thatsmight have different response mechanisms during the paralysis of CoTS.These results will provide basis for further studying the toxins’ secretionmechanism of.
5 Conclusions
In this study,sofwere identified and the sequence, structure, disulfide bonding bridge and phylo- genetic tree were analyzed.The comprehensive results showed that the fours might be different toxin genes. The expressions ofs in different predation periods of the giant triton snail () revealed that theswere highly expressed in salivary gland, digestive gland and liver of predation treatment group.
Acknowledgements
This work was supported by the National Natural Sci- ence Foundation of China (No. 42176129), the Strategic Priority Research Program of the Chinese Academy of Sci- ences (No. XDA13020206), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0402), and the Planning Project of Guang- dong Province, China, Guangdong Provincial Key Labora- tory of Applied Marine Biology (No. 2020B1212060058).
Abalde, S., Tenorio, M. J., Afonso, C. M. L., and Zardoya, R., 2018. Conotoxin diversity in(Born, 1778) and the convergent origin of piscivory in the Atlantic and Indo-Pacific cones., 10(10): 2643-2662, DOI:10.1093/gbe/evy150.
Bandel, K., 1984.. EJ Brill, Leiden, 1-199.
Biggs, J. S., Watkins, M., Puillandre, N., Ownby, J.P., Lopez-Vera, E., Christensen, S.,., 2010. Evolution ofpep-tide toxins: Analysis ofReeve, 1844., 56(1): 1-12, DOI:10.1016/j.ympev.2010.03.029.
Bose, U., Suwansa-Ard, S., Maikaeo, L., Motti, C. A., Hall, M. R.,and Cummins, S. F., 2017a. Neuropeptides encoded within a neural transcriptome of the giant triton snail, a Crown-of-Thorns Starfish predator., 98: 3-14, DOI:10.1016/j.peptides.2017.01.004.
Bose, U., Wang, T., Zhao, M., Motti, C. A., Hall, M. R., and Cum-mins, S. F., 2017b. Multiomics analysis of the giant triton snail salivary gland, a crown-of-thorns starfish predator., 7: 6000, DOI:10.1038/s41598-017-05974-x.
Brown, E., Masinde, E. L. K., and Woodcock, B. G., 2016. Ef- fects of conopeptide-containing venom from seven species ofgastropoda on the chick biventer-cervicis nerve-mus- cle assessed using the ConoServer database., 54(7): 544-554, DOI:10.5414/cp202667.
Chesher, R. H., 1969. Destruction of Pacific corals by sea star., 165(3890): 280-283, DOI:10.1126/science.165.3890.280.
Cowan, Z. L., Pratchett, M., Messmer, V., and Ling, S., 2017.Known predators of crown-of-thorns starfish (spp.) and their role in mitigating, if not preventing, population out- breaks., 9(1): 7, DOI:10.3390/d9010007.
Cui, L. B., Lu, Y. H., Liu, C. L., and Tang, H., 2000. Light and electron microscopic studies on the salivary glands ofIno., 22(1): 141-144 (in Chinese with English abstract).
Dutertre, S., Jin, A. H., Kaas, Q., Jones, A., Alewood, P. F., and Lewis, R. J., 2013. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom., 12(2): 312-329, DOI:10.1074/mcp.M112.021469.
Endean, R., 1972. Aspects of molluscan pharmacology., 7:421-466.
Garcia Ortega, L., Alegre Cebollada, J., Garcia Linares, S., Bruix, M., Martinez del Pozo, A., and Gavilanes, J. G., 2011. The be- havior of sea anemone actinoporins at the water-membrane in-terface., 1808(9): 2275-2288, DOI:10.1016/j.bbamem.2011.05.012.
Hall, A. E., and Kingsford, M. J., 2016. Variation in the popula- tion demographics ofin response to pre- dators., 35(4): 1173-1185, DOI:10.1007/s00338-016-1486-0.
Hall, M. R., Kocot, K. M., Baughman, K. W., Fernandez-Val-verde, S. L., Gauthier, M. E. A., Hatleberg, W. L.,., 2017. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest., 544(7649): 231-234, DOI:10.1038/nature22033.
Kaas, Q., Westermann, J.C., and Craik, D. J., 2010. Conopeptide characterization and classifications: An analysis using Cono-Server., 55(8): 1491-1509, DOI:10.1016/j.toxicon.2010.03.002.
Kaas, Q., Westermann, J.C., Halai, R., Wang, C. K. L., and Craik,D. J., 2008. ConoServer, a database for conopeptide sequencesand structures., 24(3): 445-446, DOI:10.1093/bioinformatics/btm596.
Kaas, Q., Yu, R., Jin, A.H., Dutertre, S., and Craik, D. J., 2012. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database., 40(D1): D325-D330, DOI:10.1093/nar/gkr886.
Kang, K. H., and Kim, J. M., 2004. The predation of trumpetshell,sp., on eight different marine invertebrate spe- cies., 35(13): 1202-1206, DOI:10.1111/j.1365-2109.2004.01124.x.
Li, Q., Barghi, N., Lu, A., Fedosov, A. E., Bandyopadhyay, P. K., Lluisma, A. O.,., 2017. Divergence of the venom exogene repertoire in two sister species of turriconus., 9(9): 2211-2225, DOI:10.1093/gbe/evx157.
Luo, S. L., Christensen, S., Zhangsun, D. T., Wu, Y., Hu, Y., Zhu, X. P.,., 2013. A novel inhibitor ofα9α10 nicotinic acety- lcholine receptors fromdelineates a new cono- toxin superfamily., 8(1): e54648, DOI:10.1371/jour-nal.pone.0054648.
Luo, S. L., Wu, Y., Zhu, X. P., and Feng, J. C., 2007. Cloning of novel O-superfamily conotoxin ofs collected from Hainan., 26(1): 1-7 (in Chinese with English abstract).
Lynch, S. S., Cheng, C. M., and Yee, J. L., 2006. Intrathecal zico- notide for refractory chronic pain., 40(7-8): 1293-1300, DOI:10.1345/aph.1G584.
McGivern, J. G., 2007. Ziconotide: A review of its pharmacology and use in the treatment of pain., 3(1): 69-85, DOI:10.2147/nedt.2007.3.1.69.
Miljanich, G. P., 2004. Ziconotide: Neuronal calcium channel blocker for treating severe chronic pain., 11(23): 3029-3040, DOI:10.2174/0929867043363884.
Peng, C., Yao, G., Gao, B. M., Fan, C. X., Bian, C., Wang, J. T.,., 2016. High-throughput identification of novel conoto- xins from the Chinese tubular cone snail () by multi-transcriptome sequencing., 5: 17, DOI:10.1186/s13742-016-0122-9.
Percharde, P. L., 1972. Observations on the gastropod,, in Trinidad and Tobago., 85: 84-92.
Phuong, M. A., Mahardika, G. N., and Alfaro, M. E., 2016. Die- tary breadth is positively correlated with venom complexity in cone snails., 17: 401, DOI:10.1186/s12864-016-2755-6.
Poulsen, A. L., 1995. Coral reef gastropods–A sustainable re- source?, 2: 142-145.
Robinson, S. D., Li, Q., Lu, A., Bandyopadhyay, P. K., Yandell, M., Olivera, B. M.,., 2017. The venom repertoire of(Chemnitz, 1777), the glory of the sea., 15(5): 145, DOI:10.3390/md15050145.
Safavi-Hemami, H., Siero, W. A., Gorasia, D. G., Young, N. D., Macmillan, D., Williamson, N. A.,, 2011. Specialisation of the venom gland proteome in predatory cone snails reveals functional diversification of the conotoxin biosynthetic path- way., 10(9): 3904-3919, DOI:10.1021/pr1012976.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S., 2013. MEGA6: Molecular evolutionary genetics analysis ver- sion 6.0., 30(12): 2725-2729.
Zhang, G. G., Xu, M., Zhang, C. L., Jia, H. X., Zhang, H., He, M. X.,., 2021. Comparative transcriptomic and expression profiles between the foot muscle and mantletissues in the gianttriton snail., 12: 632 518, DOI:10.3389/fphys.2021.632518.
Zhang, L. P., Xia, J., Peng, P. F., Li, P., Luo, P., and Hu, C. Q., 2013. Characterization of embryogenesis and early larval develop- ment in the Pacific triton,(Gastropoda: Ca- enogastropoda)., 57(3): 237-246, DOI:10.1080/07924259.2012.753472.
(August 3, 2021; revised November 22, 2021; accepted April 29, 2022)
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2023
Corresponding author. E-mail: lwg@scsio.ac.cn
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- The Subduction Structure Beneath the New Britain Island Arc and the Adjacent Region from Double-Difference Tomography
- Differences of Polygonal Faults with Irregularly Polygonal Geometries: A Case Study from the Changchang Sag of Qiongdongnan Basin, Northern South China Sea
- Characterization of Bacterial Communities in Aerosols over Northern Chinese Marginal Seas and the Northwestern Pacific Ocean in Autumn
- Assessment and Application of Beach Quality Based on Analytic Hierarchy Process in Yangkou Beach, Qingdao
- Role of Resuspended Sediments as Sources of Dissolved Inorganic Phosphorus Along Different Dimensions in the Subei Shoal, South Yellow Sea, China
- Pharmacokinetics of Enrofloxacin and Its Metabolite in Carp (Cyprinus carpio) After a Single Oral Administration in Medicated Feed
