龙岩山麻鸭蛋品质性状的全基因组关联研究
2023-03-07孙艳发吴琼林如龙陈红萍甘秋云沈玥王亚茹薛鹏飞陈飞帆刘健涛周陈鑫兰诗诗潘浩哲邓凡岳稳江宵兵李焰
孙艳发,吴琼,林如龙,陈红萍,甘秋云,沈玥,王亚茹,薛鹏飞,陈飞帆,刘健涛,周陈鑫,兰诗诗,潘浩哲,邓凡,5,岳稳,江宵兵,李焰
龙岩山麻鸭蛋品质性状的全基因组关联研究

1龙岩学院生命科学学院,福建龙岩 364012;2龙岩学院/福建省家畜传染病防治与生物技术重点实验室/预防兽医学与生物技术福建省高校重点实验室,福建龙岩 364012;3龙岩市新罗区农业局山麻鸭原种场,福建龙岩 364031;4福建省畜牧总站,福州 350003;5福建农林大学动物科学学院(蜂学学院),福州 350002
【目的】通过全基因组关联研究(genome-wide association study,GWAS)技术筛选和鉴定鸭蛋品质性状的单核苷酸多态性(single nucleotide polymorphisms,SNPs)位点及候选基因,为龙岩山麻鸭蛋品质性状分子育种提供参考。【方法】试验测定产蛋后期235只龙岩山麻鸭母鸭蛋品质性状,包括蛋重(egg weight,EW)、蛋形指数(egg shaped index,ESI)、蛋壳厚(eggshell thickness,EST)、蛋壳强度(eggshell strength,ESS)、蛋壳颜色L*、a*、b*值(eggshell colour L*, a*, b*,ESCL、ESCA和ESCB)、蛋白高度(albumin height,AH)、哈氏单位(Haugh unit,HU)、蛋黄颜色(egg yolk colour,EYC)、蛋黄重(egg yolk weight,EYW)和蛋黄比例(egg yolk percentage relative to egg weight,EYP)。使用ASReml-R 4.1软件多性状动物模型对蛋品质性状进行遗传参数估计。使用简化基因组测序技术对鸭血液基因组DNA进行SNP分型,分型后进行蛋品质性状与这些SNPs间的GWAS研究。【结果】龙岩山麻鸭蛋品质性状中,EW、ESI、EST、ESL、ESA和AU具有中高等的遗传力,遗传力在0.21—0.70之间。EW与AU存在较强的正遗传相关(g= 0.91±0.37)。ESI与EYC存在较强的遗传负相关(g= -0.98±1.03)。EST与ESS具有表型正相关(p= 0.41±0.06),与ESA具有遗传和表型负相关(g= -0.86±0.25和p= -0.15±0.07),与ESB具有遗传和表型正相关(g= 0.96±0.37和p= 0.18±0.07)。ESA与ESB具有遗传和表型负相关(g= -0.64±0.28和p= -0.31±0.06)。GWAS研究结果表明,7个SNPs位点与ESI、EST和EYC达到5%基因组水平显著关联(<4.74×10-6),涉及6个候选基因。与ESI关联的SNP(chr20:11135563:G:C)位点位于20号染色体含有75A富含亮氨酸重复序列(leucine rich repeat containing 75A)基因内。与EST关联的2个SNPs(chr13:5766560:A:G和chrZ:968819:C:T)位点分别位于13号下游6.86 kb处和Z染色体转录因子4(transcription factor 4)基因内。与EYC关联的4个SNPs位点,其中1个(chr2:38155965:G:A)位于2号染色体钾电压门控通道亚家族H成员8(potassium voltage-gated channel subfamily H member 8)基因内;3个SNPs位于9号染色体上的位点,2个(chr9:22623156:G:A和chr9:22623155:T:C)位于胰岛素受体底物1(insulin receptor substrate 1)内、1个(chr9:22490158:A:T)位于内。同时发现81个SNPs位点与蛋品质性状达到基因组水平潜在关联(<9.48×10-5)。13个与EYC关联的SNPs位点集中在9号染色体0.84 Mb(22.16—23.00 Mb)区域内。【结论】估计了龙岩山麻鸭蛋品质性状的遗传参数,通过蛋品质性状GWAS研究鉴定了影响ESI、EST和EYC性状的7个显著的SNPs位点、6个候选基因和1个候选基因区域,这些结果为龙岩山麻鸭蛋品质性状分子育种提供参考信息。
龙岩山麻鸭;蛋品质;遗传参数;单核苷酸多态性;全基因组关联研究
0 引言
【研究意义】禽蛋品质包括外部品质和内部品质。外部品质主要包括蛋重(egg weight,EW)、蛋壳颜色(eggshell color)、蛋形指数(egg shape index,ESI)、蛋壳厚度(eggshell thickness,EST)和蛋壳强度(eggshell strength,ESS),影响消费者的选择和生产效益;内部品质主要包括蛋白高度(albumen height,AH)、哈氏单位(Haugh unit,HU)、蛋黄颜色(egg yolk color,EYC)、蛋黄重(egg yolk weight,EYW)、蛋黄比例(egg yolk percentage relative to egg weight,EYP),影响蛋的新鲜程度和营养价值[1]。日粮营养水平、饲养管理、产蛋阶段以及蛋的存储时间等因素影响蛋品质。蛋品质为数量性状,受微效多基因控制[2-3]。研究人员[1,4-9]通过全基因组关联研究(genome-wide association study,GWAS)技术,鉴定了影响鸡蛋品质性状的SNP位点和候选基因,鸡蛋品质性状的分子遗传机制逐渐被揭示。目前,鸭蛋品质的候选基因和分子遗传机制尚不完全清楚。通过GWAS技术鉴定影响鸭蛋品质性状的候选基因和分子标记,可为蛋鸭蛋品质分子选育提供理论基础。【前人研究进展】王珍珍[10]采用重测序法对166只绍兴鸭血液基因组进行基因分型,通过GWAS技术检测到10个与产蛋性状显著关联的SNP,未检测到与蛋品质性状显著关联的SNP位点。LIU等[3]以352只北京鸭和麻鸭构建的F2杂交群体为研究对象,遗传参数估计结果表明蛋品质性状的遗传力在0.16—0.71之间。GWAS技术鉴定到影响AH和HU的候选区域在5号染色体5.8 Mb(14.7—20.5 Mb),该区域内111候选基因中的黏蛋白6和低密度脂蛋白受体A类结构域包含3为影响鸭蛋清组成成分的重要候选基因。【本研究切入点】山麻鸭为我国主要蛋鸭地方品种之一,原产地为福建龙岩市新罗区龙门镇,2017年中华人民共和国农业农村部正式批准对“龙岩山麻鸭”实施农产品地理标志登记保护[11]。龙岩山麻鸭具有体型小、性早熟、产蛋量高等特点,其蛋品质性状候选基因和分子遗传机制鲜见报道。龙岩山麻鸭高产系在进行选育的过程中,于72周产蛋结束后收集种蛋进行下一世代孵化。【拟解决的关键问题】以龙岩山麻鸭高产系第4世代为研究对象,采用简化基因组测序(genotyping- by-sequencing,GBS)技术[12]对龙岩山麻鸭母鸭血液基因组DNA(genome DNA,gDNA)进行SNP分型,进行产蛋末期蛋品质性状与SNPs间的GWAS,为揭示龙岩山麻鸭蛋品质性状的候选基因和分子选育提供理论基础。
1 材料与方法
1.1 试验动物
龙岩山麻鸭高产系第4世代群体于2018年2月初孵化,2018—2019年饲养于福建省龙岩市新罗区农业局山麻鸭原种场。该群体在同一时间孵化,在相同的营养与环境条件下进行单笼饲养,常规免疫。
1.2 方法
1.2.1 蛋品质测定 龙岩山麻鸭产蛋末期71—72周龄时开始收集鸭蛋,剔除破蛋、软壳蛋和双黄蛋,称重,记为EW。蛋品质测定于龙岩学院生命科学学院实验室进行。使用游标卡尺(日本Mitutoyo 公司)测定鸭蛋的长短径,计算ESI。使用蛋壳厚度计(TQ-1A,南京铭奥仪器设备有限公司)测定蛋壳钝端、锐端和中部位置的蛋壳厚度,平均值作为EST。使用蛋壳强度测定仪(KQ-1A,北京天翔飞域科技有限公司)测定ESS。采用WSC-S色差计(上海申光有限公司)以CIELAB体系测定蛋壳颜色。CIELAB体系测定结果记录为蛋壳颜色的亮度值(L*,ESCL)、红色值(a*,ESCA)和黄色值(b*,ESCB)。使用蛋黄分离器分离出蛋黄,称重记EYW,并计算EYP。使用罗氏蛋黄比色扇(日本Robotmation公司)测定蛋黄颜色。使用蛋白高度测定计(日本Mitutoyo 公司)测定蛋白两个位置的AU,计算平均值。根据下列公式计算HU:
=100×10(-1.7×0.37+7.57)
其中,HU为哈氏单位,AU为蛋白高度(mm),EW为蛋重(g)。
1.2.2 测序分型 72周末期进行龙岩山麻鸭翅静脉采血,柠檬酸钠(ACD)抗凝,液氮冷冻后-80℃冰箱保存备用。酚氯仿法提取血液gDNA。采用内切酶对gDNA 进行酶切,构建文库后进行测序。GBS文库构建的方法为:用I限制性内切酶对gDNA进行酶切。酶切后的片段两端加Solexa P1、P2接头(adapter)。加接头后,使用III和III限制性内切酶组合再次进行酶切。使用PCR扩增两端分别含有P1和P2接头的序列,形成DNA片段池(pooling),电泳回收所需区间的DNA片段。AMPure XP beads试剂盒(美国Beckman公司)纯化上述 PCR 产物,获得GBS文库。文库质检合格后使用Illumina公司NovaseqTM测序平台进行双末端(Paired-End)150测序。测序得到的原始数据(raw data)经过过滤得到高质量的clean data。Clean data通过BWA软件[13]对比到鸭基因组(IASCAAS_PekingDuck_PBH1.5,GCF_003850225.1)[14]上,比对结果经SAMTOOLS软件[15]去除重复。采用SAMTOOLS软件(参数为-q 1 -C 50 -t AD,DP -m 2 -F 0.002)的进行多个样本SNP的检测,得到每个样本的SNP分型数据。
1.2.3 测序数据的质控 使用Plink V1.9软件[16]进行测序后基因型数据的质量控制。选择标准设置为:个体基因型的缺失率小于20%;SNP位点缺失率小于10%;最小等位基因频率大于5%;哈迪温伯格平衡的P值大于1×10-6。剔除不符合上述条件的样本和SNP位点。
1.3 数据处理与统计分析
1.3.1 表型值的描述性统计 使用Minitab V17.0软件(美国Minitab Inc)对表型数据进行描述性统计分析,剔除异常值,计算蛋品质性状的平均数(mean)、最小值(Min)、最大值(Max)、标准差(standard deviation,SD)和变异系数(coefficient of variation,CV)。
1.3.2 遗传参数估计 使用VSN国际有限公司的 ASReml-R 4.1软件的多性状动物模型对蛋品质性状的遗传参数(遗传力和遗传相关)相关进行估计,并计算表型相关。模型如下:
=++
式中,为蛋品质性状的表型值向量,为固定效应的向量(包括总体均值),为加性遗传效应的向量,为随机误差的向量,、分别为固定效应、加性遗传效应的指定矩阵。
结果以“平均数±标准误”的形式表示,采用似然比检验(likelihood-ratio test,LRT)法对遗传相关和表型相关进行显著性检验,<0.05表明性状具有相关性。
1.3.3 群体结构分析 使用 Plink V1.9软件[16]中IBS距离聚类法检测试验群体是否存在分层现象。该过程以25个SNPs为一个窗口,5个SNPs为步移,利用indep-pairwise命令计算窗口内标记成对的r2值,阈值设为 0.2,筛选所有常染色体上独立的SNPs 标记[17]。利用这些独立标记使用Plink V1.9软件主成分(principal component,PC)分析程序计算每个样本的PC,并计算每个PC解释群体结构变异的百分比。使用R V4.0.4软件[18]以主成分1(PC1)和2(PC2)绘制群体结构图。
1.3.4 关联研究 使用Plink 1.9软件中的线性回归模型进行蛋品质性状的GWAS。为了消除群体结构对关联分析结果的影响,以前10个PC为协变量。使用模型为:
=+++
式中,为表型性状值,为总体均值,为主成分效应,为SNP效应,为随机残差。
为了减少多重检验带来的假阳性,以连锁不平衡(linkage disequilibrium,LD)修正的Bonferroni方法[19]对GWAS结果的值进行校正。独立检验数为群体结构分析中获得的独立SNP数量[17],全基因组显著和潜在关联阈值的计算公式为:
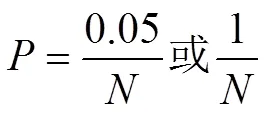
式中,为Bonferroni校正的全基因组显著或潜在关联的值,为群体结构分析中获得的独立SNPs数量。
2 结果
2.1 表型值的描述性统计
蛋品质性状表型值的描述性统计见表1。龙岩山麻鸭蛋品质性状中EST、ESS、ESCA、ESCB、HU、EYC、EYW、EYP的CV范围在10%—50%之间,表型性状分离明显,有助于基因定位。其中ESCA为负值,说明本研究使用群体中龙岩山麻鸭蛋壳颜色均偏绿色。
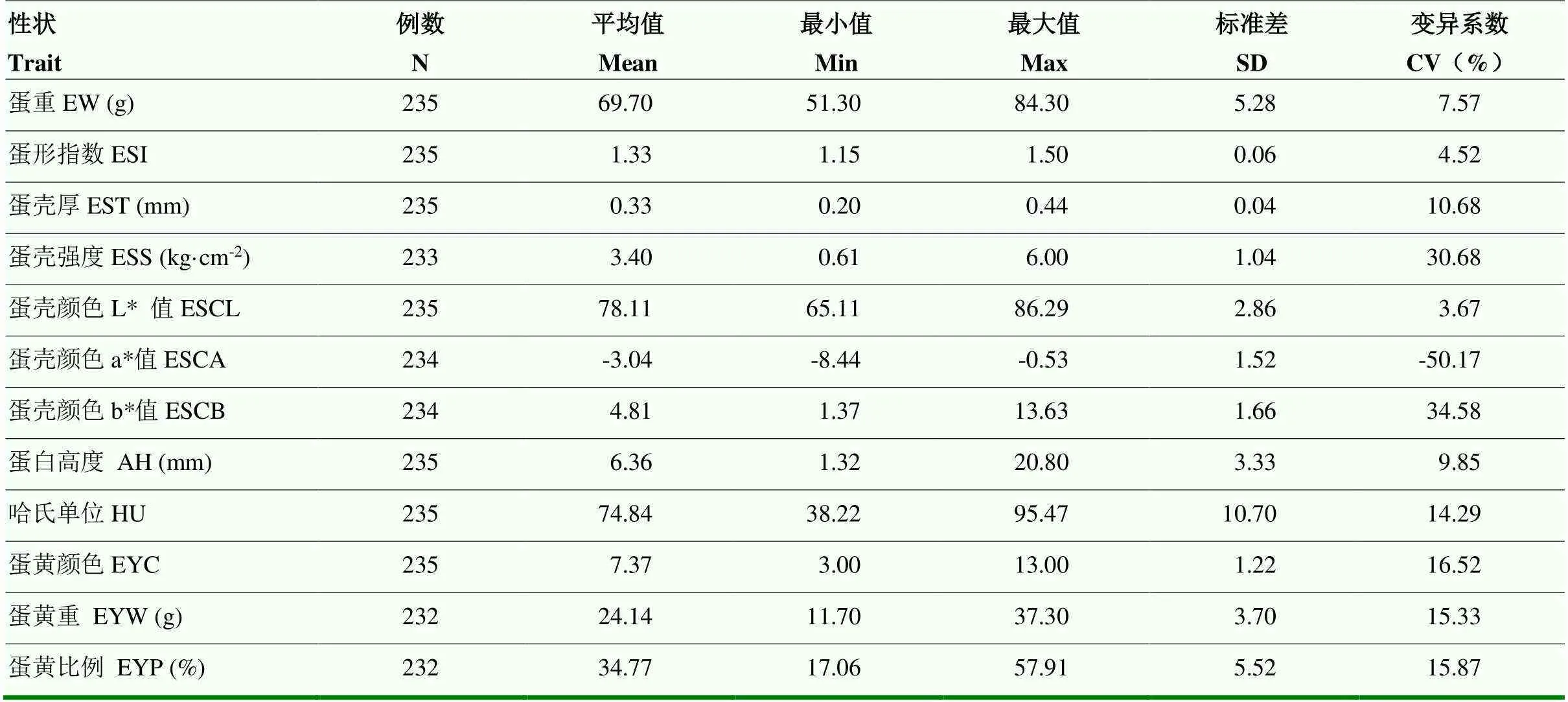
表1 蛋品质性状表型值的描述性统计
2.2 遗传参数估计
蛋品质性状的遗传参数估计结果见表2。EW、ESI、EST、ESL、ESA和AU遗传力在0.21—0.70之间,具有中高等遗传力;ESS、ESB、H U、EYC、EYW和EYP遗传力在0.01—0.16之间,具有较低的遗传力;其中ESA遗传力最高(2= 0.70±0.20),EYW为遗传力最低(2= 0.01±0.12)。EW与AU存在较强的正遗传相关(g= 0.91±0.37)。EW与AH、HU、EYC、EYW和EYP存在表型正相关或负相关,其中与EYW(p= 0.24±0.06)和EYP(p= -0.26±0.06)相关性最大。ESI与EYC存在较强的遗传负相关(g= -0.98±1.03)和较弱的表型正相关(p= 0.07±0.06)。EST与ESS具有表型正相关(p= 0.41±0.06),与ESA具有遗传和表型负相关(g= -0.86±0.25,p= -0.15±0.07),与ESB具有遗传和表型正相关(g= 0.96±0.37,p= 0.18±0.07)。ESA与ESB具有遗传和表型负相关(g= -0.64±0.28,p= -0.31±0.06)。EYW和EYP具有较强的表型正相关(p= 0.87±0.02)。
上三角为遗传相关,下三角为表型相关;“-”表明性状间没有相关性; *<0.05;**<0.01;***<0.001
Genetic correlations are given above the diagonal and phenotypic correlation below the diagonal; “-” indicates that there was no correlation among traits
2.3 基因型数据质控结果
基因型数据经过质量控制后,303个个体和62 706 SNP用于后续的分析。SNPs在各染色体上的分布数量情况见表3。
2.4 群体结构分析
常染色体上的SNP经筛选后,共得到10 428个独立SNP标记(表3)。由群体结构主成分分析图(图1)可知,303个龙岩山麻鸭群体明显分成几个簇,存在分层现象,容易造成GWAS结果中出现假阳性和假阴性。主成分分析结果中前10个PC解释了92.54%群体结构变异。因此,本研究以前10个PC为协变量,以消除群体分层对关联分析结果的影响[20]。
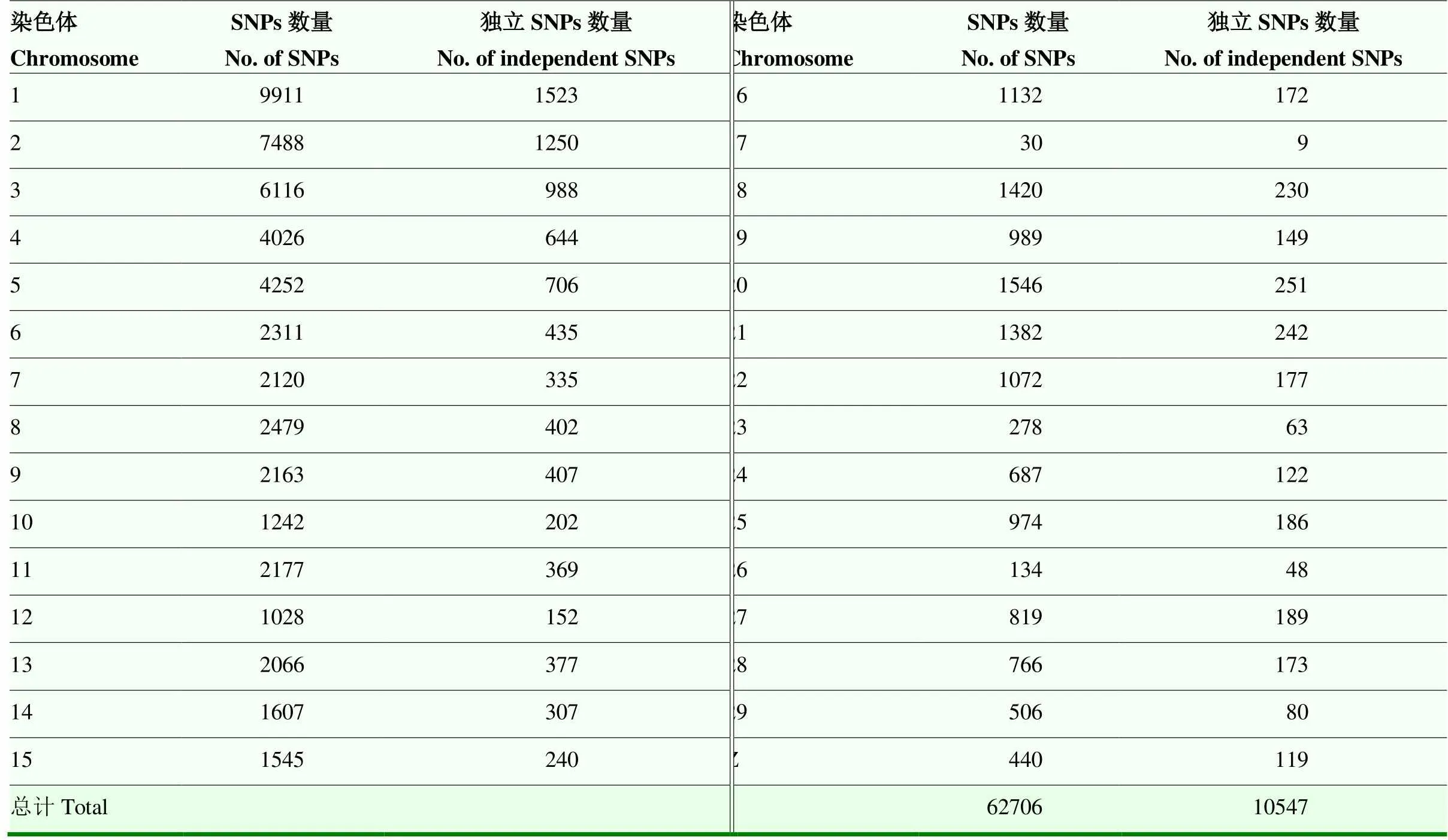
表3 质控后和独立的SNPs标记在各条染色体上的分布
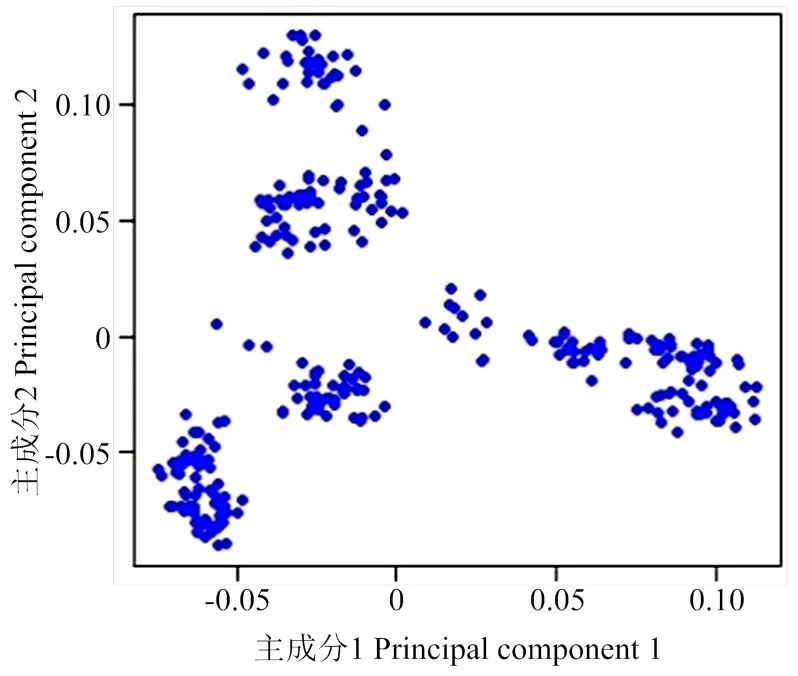
图1 群体结构主成分分析图
2.5 关联研究结果
由于基因组中独立SNP标记数量为10 547(表3),Bonferroni校正的5%基因组显著水平值阈值为4.74×10-6(0.05/10 547),基因组潜在关联水平的阈值为9.48×10-5(1/10 547)。本研究发现7个SNP位点与ESI、EST和YC达到5%基因组显著关联(<4.74×10-6)(表4和图2—4),81个SNP位点与蛋品质性状达到基因组水平潜在关联(<9.48×10-5)(表5)。
2.5.1 蛋形指数 1个SNP(chr20:11135563:G:C与)ESI达到5%Bonferroni校正的基因组显著关联(= 14.83×10-6)(表4和图2)。该SNP位点位于20号染色体含有75A富含亮氨酸重复序列(leucine rich repeat containing 75A,)基因内。此外,10个SNP位点与ESI达到基因组潜在关联(<9.48× 10-5)(表5和图2)。其中3个SNPs(chr3:81386471: A:G、chr5:7009743:G:A和chr25:6315960:T:C)分别位于3号、5号和25号染色体上,2个SNP(chr16: 2579629:A:T和chr16:3098786:G:A)位于16号染色体上,位于5'-核苷酸酶ecto(5'-nucleotidase ecto,)、细胞周期素依赖激酶样1(cyclin dependent kinase like 1,)、和内或下游;5个SNPs(chr15:11227397:T:C,chr15: 11227409: G:A,chr15:13294556:C:T,chr15:13395347: A:G和chr15:14536844:G:A)位于15号染色体3.31 Mb(11.23—14.54 Mb)区域内。
2.5.2 蛋壳厚度 2个SNP(chr13:5766560:A:G和chrZ:968819:C:T)与EST达到Bonferroni校正5%基因组水平显著关联(= 1.36×10-6,1.96×10-6)(表4和图3)。SNP chr13:5766560:A:G位于13号下游6.86 kb处。SNP chrZ:968819:C:T位于Z染色体转录因子4(transcription factor 4,)基因内。此外,9个SNP与EST达到Bonferroni校正的基因组潜在关联(<9.48×10-5),包括2号染色体上锌指蛋白804B(zinc finger protein 804B,)基因内的3个SNP(chr2:22513511: C:A、chr2:22582514:A:G和chr2:22582670:G:A)、6号染色体上钾双孔结构域通道亚家族K成员18(potassium two pore domain channel subfamily K member 18,)基因下游18.98 kb处和SH3 and PX结构域2A(SH3 and PX domains 2A,)基因内的2个SNP(chr6:7192663:G:A和chr6:12756206:T: A)、12号染色体凝血酶反应蛋白1型结构域包含4(thrombospondin type 1 domain containing 4,)基因内的2个SNP(chr12:140915:T:A和chr12:198263:G:A)、13号染色体基因内的1个SNP(chr13:5721234:C:T)以及19号染色体基因内的1个SNP(chr19: 4873329:A:G)。
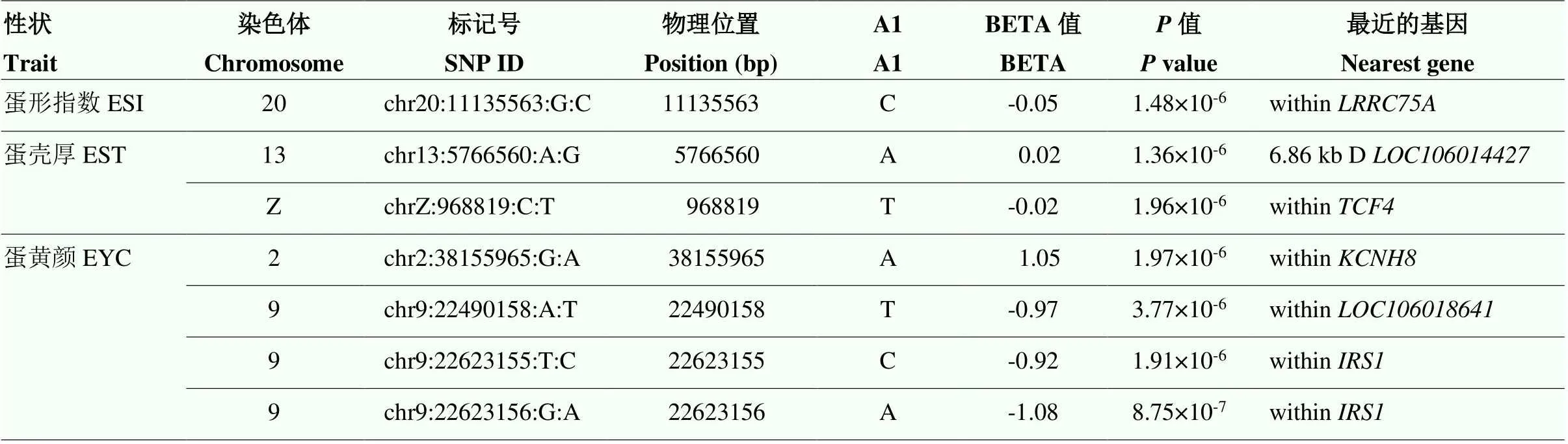
表4 Bonferroni校正5%基因组显著的SNP位点
A1,次要的等位基因;BETA,回归系数,正值表示次要等位基因提高性状值;D代表SNP位于基因的下游;within代表SNP位于基因内。下同
A1, minor allele; BETA, regression coefficient, a positive regression coefficient means that the minor allele increases traits mean; D means SNP downstream of the nearest gene; within represent SNP located within genes.The same as below
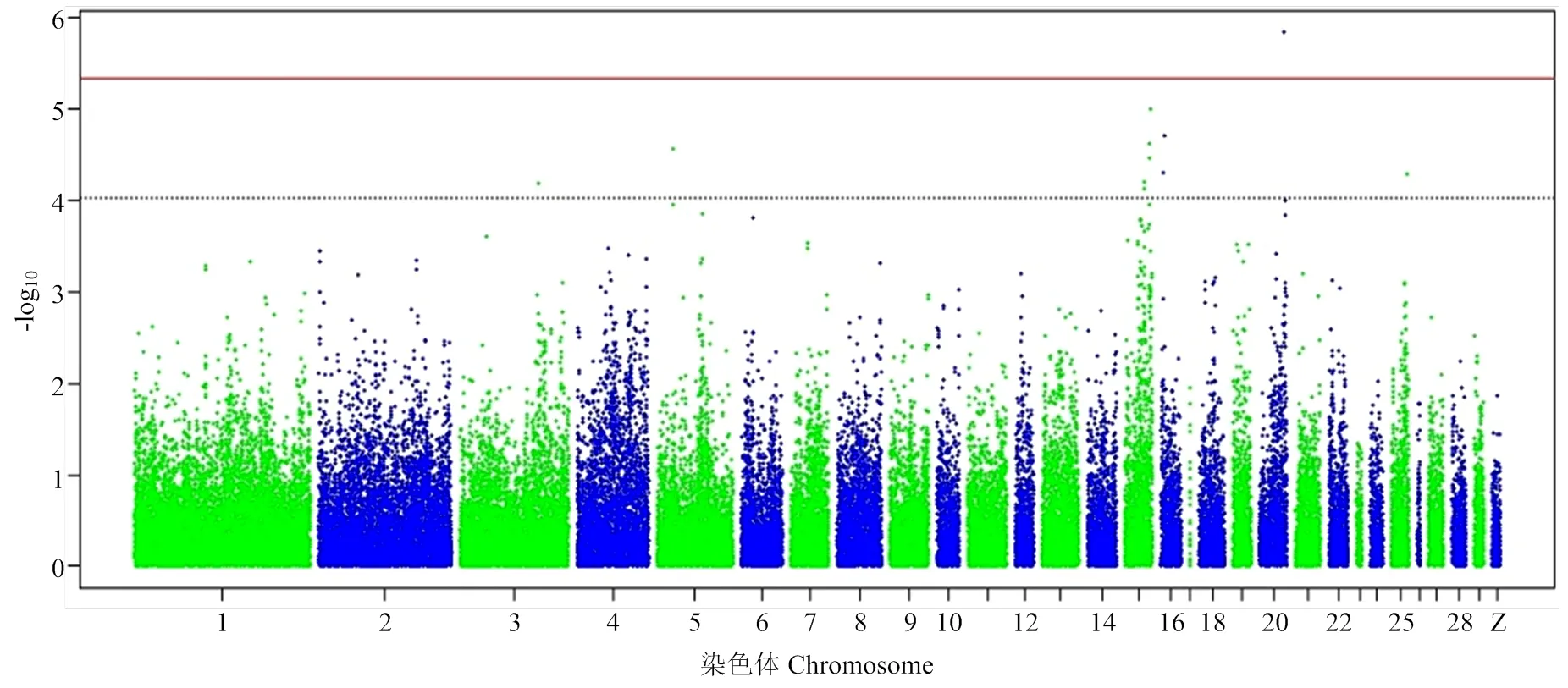
横坐标为SNPs标记在基因组中的物理位置,纵坐标为关联研究中P值的-log10转化结果。每一个点代表一个SNP标记。红色实线为达到5%全基因组显著的阈值线(-log10 (4.74×10-6)),黑色虚线为达到全基因组潜在关联的阈值线(-log10 (9.48×10-5))。下同
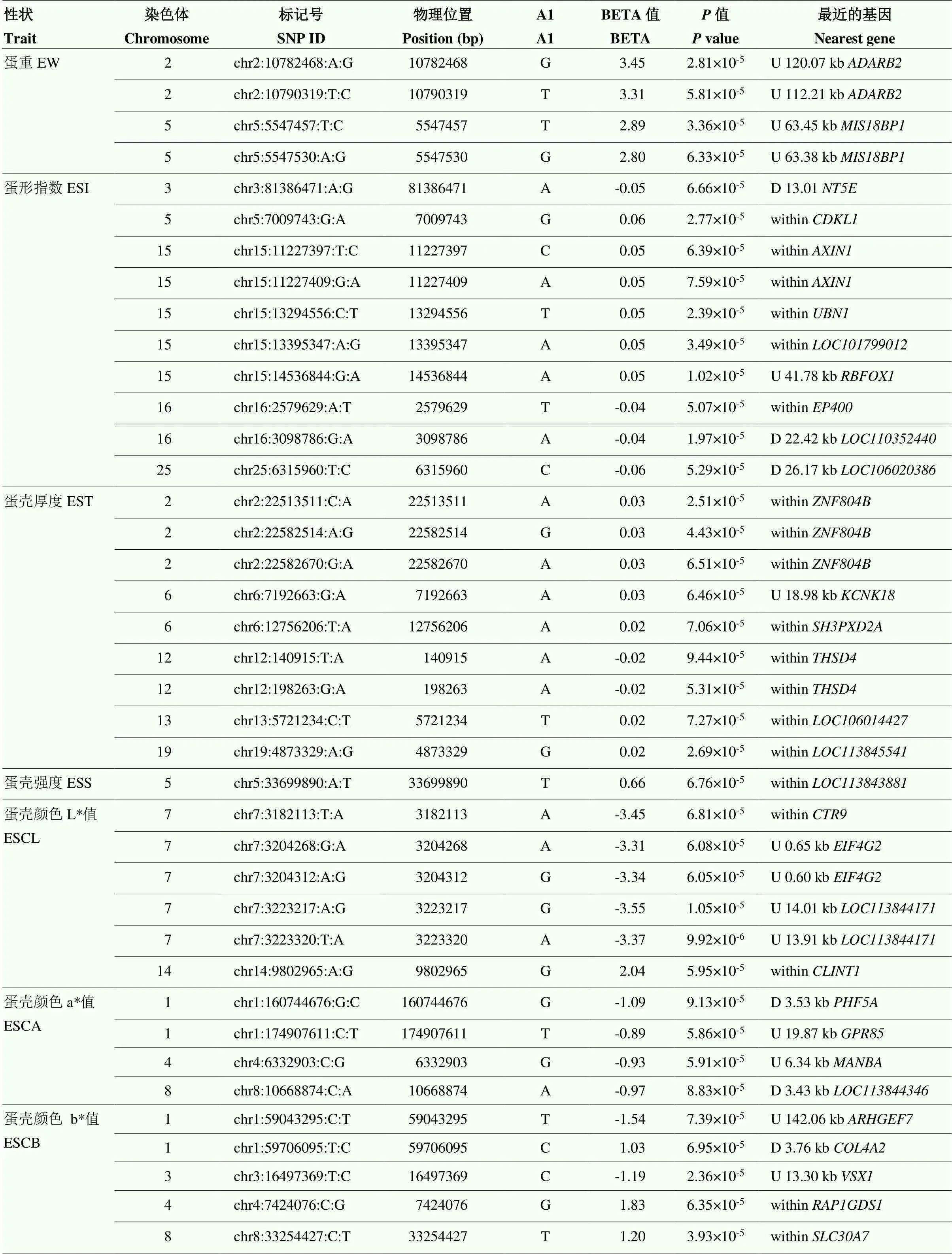
表5 基因组水平潜在关联的SNP位点
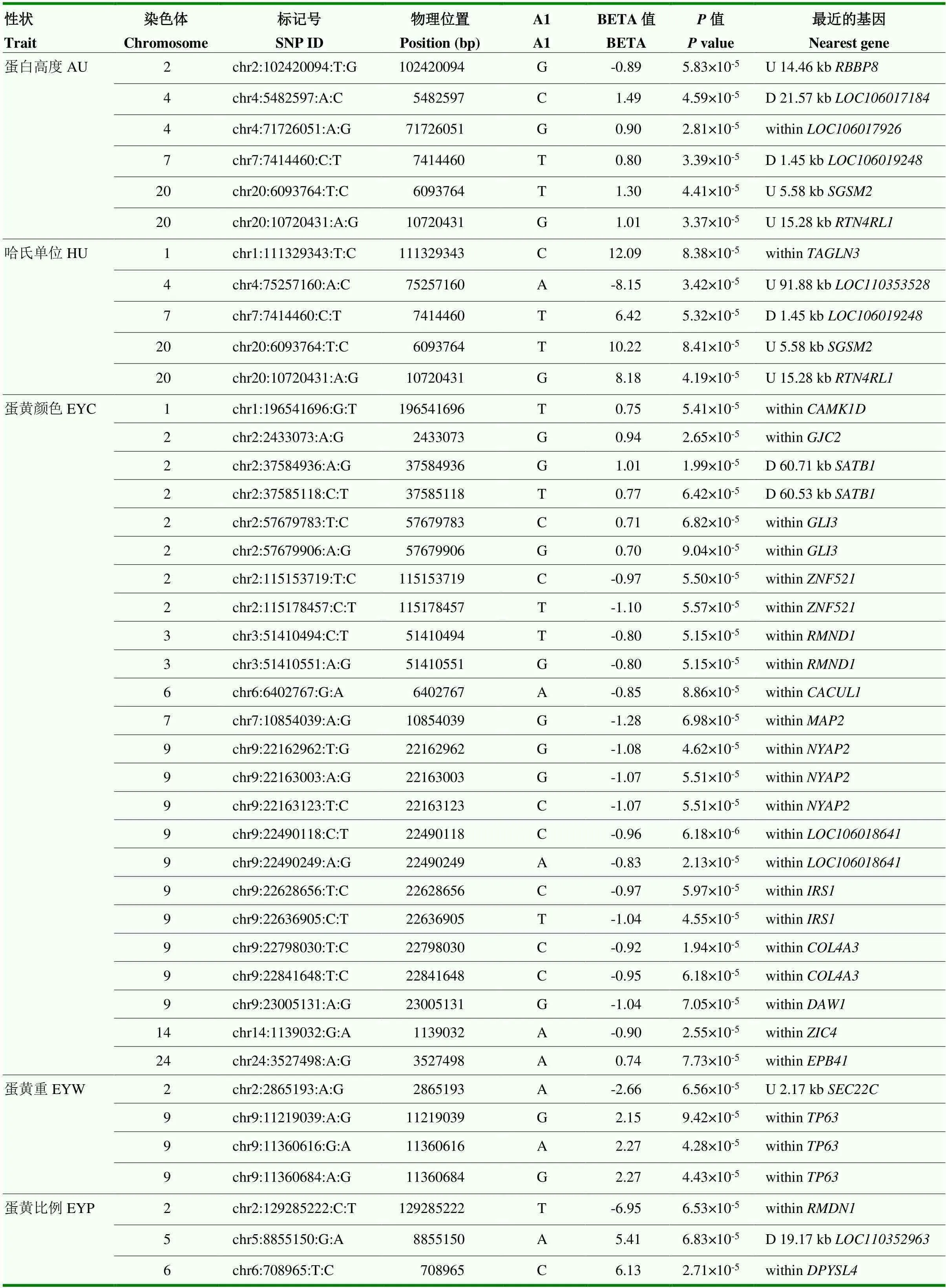
续表5 Continued table 5
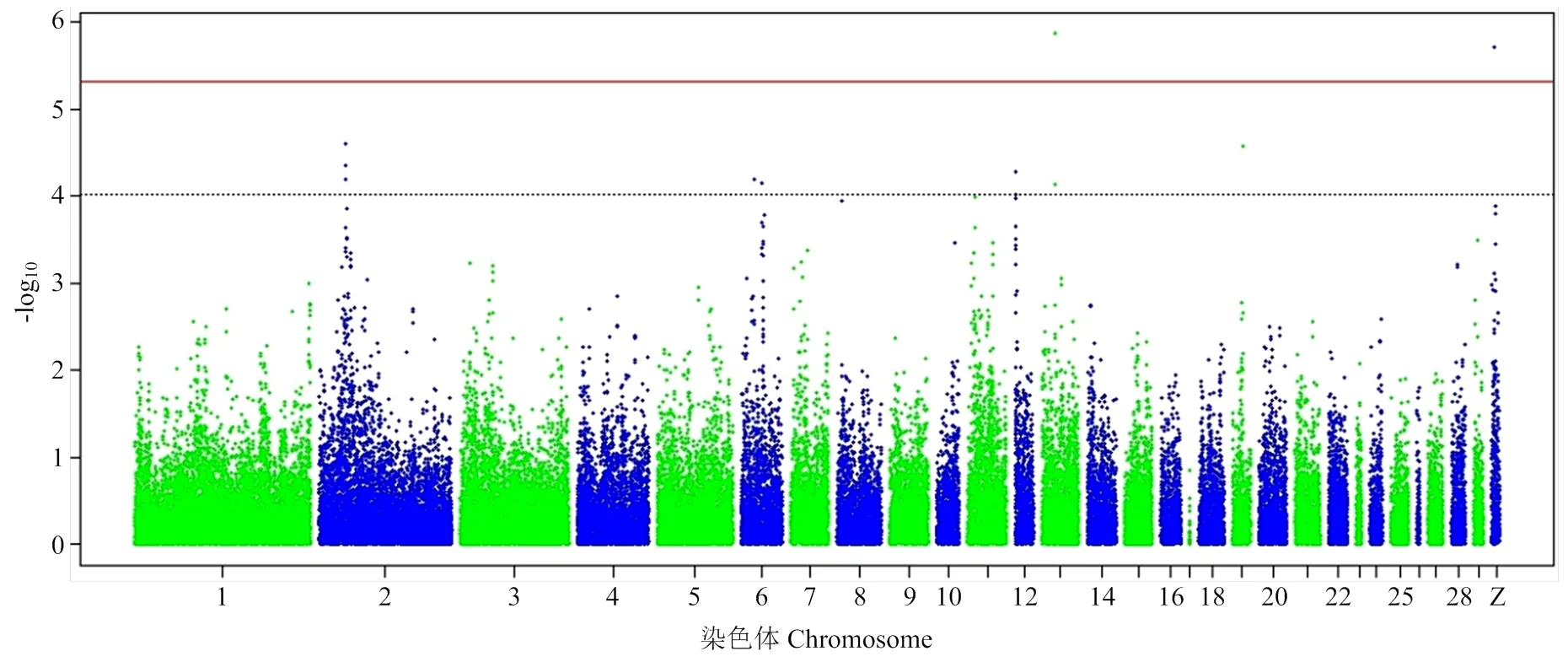
图3 蛋壳厚度全基因组关联研究曼哈顿图
2.5.3 蛋黄颜色 4个SNP(chr2:38155965:G:A,chr9:22490158:A:T,chr9:22623155:T:C和chr9: 22623156:G:A)与EYC达到Bonferroni校正5%基因组水平显著关联(<4.74×10-6)(表4和图4)。其中SNP chr2:38155965:G:A位于2号染色体钾电压门控通道亚家族H成员8(potassium voltage-gated channel subfamily H member 8,)基因内。3个SNP位于9号染色体上,2个(chr9:22623156:G:A和chr9:22623155:T:C)位于胰岛素受体底物1(insulin receptor substrate 1,)内、1个(chr9: 22490158:A:T)位于内。此外,20个SNP与EYC达到基因组水平潜在关联(<9.48×10-5)(表5),其中有10个位于9号染色体上。9号染色体上一共有13个与EYC关联的SNP集中在0.84 Mb(22.16—23.00 Mb)区域内。这一区域除和外,还包括神经元酪氨酸磷酸化磷酸肌醇 3-激酶接头2(neuronal tyrosine- phosphorylated phosphoinositide-3-kinase adaptor 2,)、Ⅳ型胶原α3链(collagen type IV alpha 3 chain,)、动力蛋白组装因子含WD重复1(dynein assembly factor with WD repeats 1,)。同时发现与EW、ESCL、ESCA、ESCB、AH、HU、EYW和EYP性状达到基因组潜在关联(<9.48× 10-5)的SNP位点总结于表5。
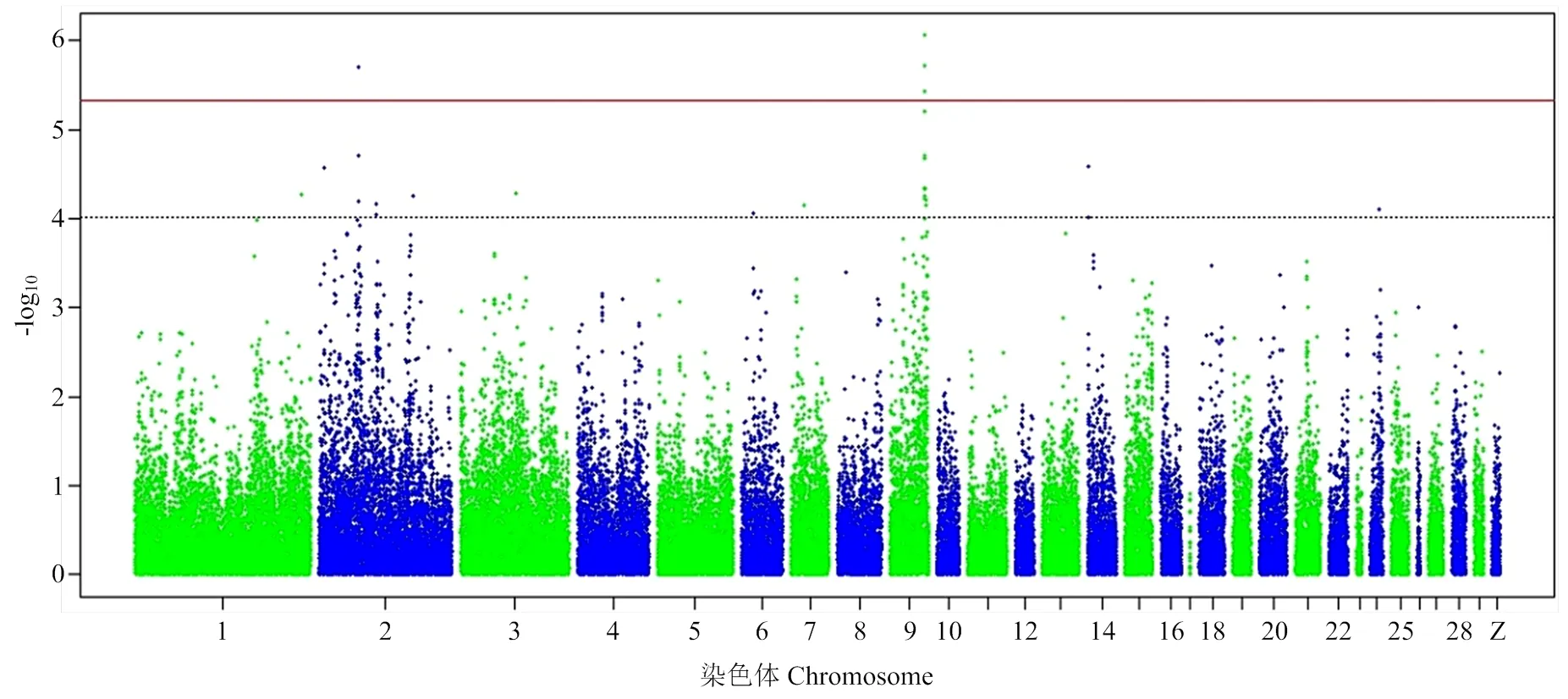
图4 蛋黄颜色全基因组关联研究曼哈顿图
3 讨论
全基因组关联研究,又称全基因组关联分析,使用高通量的基因分型技术在基因组范围内寻找SNPs与畜禽重要经济性状的关联,为鉴定畜禽重要经济性状分子标记和候选基因的一种有效方法[21]。鸭的重要经济性状GWAS研究主要集中在生长和饲养[22]、体重和胴体[23]、脂肪沉积和肉品质[24]、骨质量和饲喂效率[25]、血液成分[26]、摄食行为[27]以及肌纤维直径[28]等性状上,蛋品质性状的GWAS报道较少[3]。本研究对龙岩山麻鸭产蛋末期产蛋性状的遗传参数进行了估计,鉴定了影响产蛋性状的候选基因和候选区域,为揭示龙岩山麻鸭产蛋性状分子遗传机制和分子选育奠定了理论基础。
3.1 蛋形指数
ESI是描述蛋壳形状的经典几何参数[29],主要受家禽的产蛋率、产蛋间隔时间和蛋壳形成时物质需要量的影响[30]。本研究中龙岩山麻鸭高产系第4世代ESI遗传力为0.29±0.17,与其早期选育的ESI遗传力(2=0.34)[31]相近。ESI与蛋重、蛋白指数、哈氏单位等蛋品质性状呈表型正相关[32]。本研究中发现ESI与蛋壳颜色(ESCA)呈表型负相关,与蛋黄颜色呈遗传负相关和表型正相关,可能是由于蛋的形状与蛋壳颜色、蛋黄颜色受相近的遗传因素影响[33]。与ESI显著关联的1个SNP(chr20:11135563:G: C)位于20号染色体内。该基因编码的蛋白质可作为细胞膜上的受体[34]。研究表明,反义长链非编码RNA(lncRNA)在细胞增殖、迁移和浸润中具有重要作用[35-36]。10个与ESI达到潜在关联的位点中,5个SNP位于两个蛋白质编码基因(和)和非编码RNA(和)基因内部或下游。编码的蛋白质是一种质膜蛋白,催化细胞外核苷酸到膜透核苷的转化,该编码蛋白被用作淋巴细胞分化的决定因素[37]。编码细胞周期素依赖激酶样1蛋白,是细胞周期素依赖激酶超家族中的一员。研究发现该基因在胃癌细胞增殖和存活中起着重要的调节作用[38]。此外,研究还发现5个SNP集中在15号染色体3.31 Mb(11.23—14.54 Mb)区域内。前人的研究发现该区域内37 345 836 bp处的SNP与鸭饲料转化效率显著相关[39]。该区域内有119个基因,需要进一步精细定位研究该区域与ESI的关系。
3.2 蛋壳厚度
蛋壳对家禽产业具有重要的生物学和经济意义,EST影响商品蛋的破损率和种蛋的孵化率[40]。鸭蛋大多被加工成皮蛋或咸蛋,蛋壳在这一过程中起着至关重要的作用[41]。蛋壳矿化作用与EST密切相关,其中蛋壳特异性矩阵蛋白ovocleidins(OC-17和OC-116)、ovocalyxins(OCX-32和OCX-36)以及钙离子结合蛋白REG4发挥关键作用[42]。本研究中龙岩山麻鸭EST为中等遗传力为(2= 0.41±0.17),比早期选育遗传力(2=0.28)高[31]。本研究鉴定2个SNP(chr13:5766560:A:G和chrZ:968819:C:T)与EST显著关联。其中SNP chrZ:968819:C:T位于Z染色体内部,编码转录因子4蛋白,能够调节几种不同细胞类型的分化,在神经系统发育[41]、卵泡发育中起重要作用[43]。此外,与EST达到基因组潜在关联的位点位于蛋白质编码基因、、和内部或附近。编码锌指蛋白804B,是一种含锌指蛋白结构域的转录因子,该基因的突变与神经系统疾病有关[44]。编码钾通道蛋白超家族的一个成员,包含两个形成孔的P结构域,作为一个外向整流钾通道,与细胞电兴奋性的控制有关[45]。编码蛋白为Tks5,是一种支架蛋白和Src底物,通过其在侵袭体形成和功能中的重要作用参与细胞迁移和基质降解[46]。编码血栓反应蛋白1型结构域包含4,是一种微纤维相关蛋白,可直接与原纤蛋白-1结合并促进原纤蛋白-1基质组装[47]。上述基因在先前禽蛋蛋壳形成的多组学研究中并未提及[42]。这些基因中特别是,可能是影响鸭蛋壳厚度的新基因,需要进一步研究。
3.3 蛋黄颜色
由于消费者将蛋黄颜色与蛋所含的营养联系在一起,蛋黄颜色作为蛋品质性状中重要经济性状之一[2]。蛋黄颜色受遗传因素、养殖方式、饲料中脂质和抗氧化物质的含量等方面的影响[48]。本研究中龙岩山麻鸭蛋黄颜色遗传力低遗传力(2= 0.07±0.14),常规育种方法蛋黄颜色的遗传改良进展缓慢。因此,通过分子标记辅助育种,有助于加速龙岩山麻鸭蛋黄颜色的育种进程,提高育种效率。本研究发现9号染色体0.84 Mb(22.16—23.00 Mb)区域内13个SNP与EYC关联。该区域内有4个蛋白质编码基因包括、、和。编码胰岛素受体底物1,为胰岛素受体酪氨酸激酶的关键靶蛋白,是激素调控代谢所必需的蛋白质[49]。研究表明直接参与卵泡生长的卵巢衰老和活化[50],其突变与女性多囊卵巢综合征有关[51]。编码神经元酪氨酸磷酸化磷酸肌醇3-激酶接头2,参与神经元发育,并与 WAVE1 蛋白相互作用,参与细胞骨架建模有关[52]。编码Ⅳ型胶原α3链蛋白,参与卵黄周隙内外亚层的组成,与蛋的受精、早期胚胎发育和抗菌素防御与胚胎发生有关[53]。编码动力蛋白组装因子含WD重复1蛋白,作为外部动力蛋白臂组件,是地中海真涡虫()纤毛运动功能所必需的[54]。1个与EYC显著关联的SNP位点位于2号染色体内。该基因编码钾电压门控通道子家族H成员8,为人类Elk K+通道基因家族成员。它们的多种已知功能包括调节神经递质释放、心率、胰岛素分泌、神经元兴奋性、上皮电解质运输和平滑肌收缩[55]。上述基因和区域与EYC关系,需要进行进一步研究证实。
本研究未发现与蛋重、蛋壳颜色、蛋白高度与哈氏单位、蛋黄重与蛋白比例全基因组显著的位点,需要增加群体数量和SNP标记密度提供统计功效(statistical power),以提高对这些性状显著位点的检出[56-57]。
4 结论
本研究采用GBS基因分型技术,通过全基因组关联研究鉴定影响龙岩山麻鸭产蛋后期蛋品质性状的SNP位点88个,其中与蛋形指数、蛋壳厚和蛋黄颜色关联的7个位点(chr20:11135563:G:C、chr13:5766560:A:G、chrZ:968819:C:T、chr2:38155965:G:A、chr9:22490158: A:T、chr9:22623155:T:C和chr9:22623156:G:A)达到Bonferroni校正5%基因组显著水平,找到了、、、等候选基因。发现9号染色体上0.84 Mb(22.16—23.00 Mb)区域可能是影响龙岩山麻鸭蛋黄颜色的候选区域。本研究为揭示龙岩山麻鸭蛋品质性状的分子遗传机制,为进一步分子标记辅助选择提供了理论基础。
[1] LIU Z, SUN C J, YAN Y Y, LI G Q, SHI F Y, WU G Q, LIU A Q, YANG N. Genetic variations for egg quality of chickens at late laying period revealed by genome-wide association study. Scientific Reports, 2018, 8: 10832. doi:10.1038/s41598-018-29162-7.
[2] GAO G, GAO D, ZHAO X, XU S, ZHANG K, WU R, YIN C, LI J, XIE Y, HU S, WANG Q. Genome-wide association study-based identification of SNPs and haplotypes associated with goose reproductive performance and egg quality. Front Genet, 2021, 12: 602583. doi:10.3389/fgene.2021.602583.
[3] LIU H, ZHOU Z, HU J, GUO Z, XU Y, LI Y, WANG L, FAN W, LIANG S, LIU D, ZHANG Y, XIE M, TANG J, HUANG W, ZHANG Q, HOU S. Genetic variations for egg internal quality of ducks revealed by genome-wide association study. Animal Genetics, 2021, 52(4): 536-541. doi:10.1111/age.13063.
[4] LIU W, LI D, LIU J, CHEN S, QU L, ZHENG J, XU G, YANG N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PLoS ONE, 2011, 6(12): e28600. doi:10.1371/journal.pone.0028600.
[5] WOLC A, ARANGO J, JANKOWSKI T, DUNN I, SETTAR P, FULTON J E, O'SULLIVAN N P, PREISINGER R, FERNANDO R L, GARRICK D J, DEKKERS J C. Genome-wide association study for egg production and quality in layer chickens. Journal of Animal Breeding and Genetics, 2014, 131(3): 173-182. doi:10.1111/jbg. 12086.
[6] ZHANG G X, FAN Q C, WANG J Y, ZHANG T, XUE Q, SHI H Q. Genome-wide association study on reproductive traits in Jinghai Yellow Chicken. Animal Reproduction Science, 2015, 163: 30-34. doi:10.1016/j.anireprosci.2015.09.011.
[7] SUN C, QU L, YI G, YUAN J, DUAN Z, SHEN M, QU L, XU G, WANG K, YANG N. Genome-wide association study revealed a promising region and candidate genes for eggshell quality in an F2resource population. BMC Genomics, 2015, 16: 565. doi:10.1186/ s12864-015-1795-7.
[8] LIAO R, ZHANG X, CHEN Q, WANG Z, WANG Q, YANG C, PAN Y. Genome-wide association study reveals novel variants for growth and egg traits in Dongxiang blue-shelled and White Leghorn chickens. Anim Genet, 2016, 47(5): 588-596. doi:10.1111/age.12456.
[9] QU L, SHEN M, GUO J, WANG X, DOU T, HU Y, LI Y, MA M, WANG K, LIU H. Identification of potential genomic regions and candidate genes for egg albumen quality by a genome-wide association study. Archives Animal Breeding, 2019, 62(1): 113-123. doi:10.5194/aab-62-113-2019.
[10] 王珍珍. 不同蛋鸭品种产蛋性能的比较分析及绍兴鸭产蛋性能的全基因组关联分析[D]. 金华: 浙江师范大学, 2020.
WANG Z Z. Analysis on egg quality traits of four laying duck breeds and genome-wide association study of laying performance in Shaoxing duck[D]. Jinhua: Zhejiang Normal University, 2020. (in Chinese)
[11] 孙艳发, 李焰, 林如龙, 陈红萍, 吴琼, 李建磊, 陈羽, 林泽. 龙岩山麻鸭产蛋量和蛋重性状的遗传参数估计. 中国畜牧杂志, 2020, 56(10): 51-55. doi:10.19556/j.0258-7033.20191022-03.
SUN Y F, LI Y, LIN R L, CHEN H P, WU Q, LI J L, CHEN Y, LIN Z. Estimation of genetic parameters for egg production and weight traits in Longyan Shan-ma duck. Chinese Journal of Animal Science, 2020, 56(10): 51-55. doi:10.19556/j.0258-7033.20191022-03. (in Chinese)
[12] ROWAN B A, SEYMOUR D K, CHAE E, LUNDBERG D S, WEIGEL D. Methods for genotyping-by-sequencing. Methods in Molecular Biology (Clifton, N J), 2017, 1492: 221-242. doi:10.1007/ 978-1-4939-6442-0_16.
[13] LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009, 25(14): 1754-1760. doi:10.1093/bioinformatics/btp324.
[14] HUANG Y H, LI Y R, BURT D W, CHEN H L, ZHANG Y, QIAN W B, KIM H, GAN S Q, ZHAO Y Q, LI J W, YI K, FENG H P, ZHU P Y, LI B, LIU Q Y, FAIRLEY S, MAGOR K E, DU Z L, HU X X, GOODMAN L, TAFER H, VIGNAL A, LEE T, KIM K W, SHENG Z Y, AN Y, SEARLE S, HERRERO J, GROENEN M A M, CROOIJMANS R P M A, FARAUT T, CAI Q L, WEBSTER R G, ALDRIDGE J R, WARREN W C, BARTSCHAT S, KEHR S, MARZ M, STADLER P F, SMITH J, KRAUS R H S, ZHAO Y F, REN L M, FEI J, MORISSON M, KAISER P, GRIFFIN D K, RAO M, PITEL F, WANG J, LI N. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nature Genetics, 2013, 45(7): 776-783. doi:10.1038/ng.2657.
[15] LI H, HANDSAKER B, WYSOKER A, FENNELL T, RUAN J, HOMER N, MARTH G, ABECASIS G, DURBIN R. 1000 GENOME PROJECT DATA PROCESSING SUBGROUP. The sequence alignment/map format and SAMtools. Microbiology Spectrum, 2009, 25(16): 2078-2079. doi:10.1093/bioinformatics/btp352.
[16] PURCELL S, NEALE B, TODD-BROWN K, THOMAS L, FERREIRA M A, BENDER D, MALLER J, SKLAR P, DE BAKKER P I, DALY M J, SHAM P C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Biological Psychiatry, 2007, 81(3): 559-575. doi:10.1086/519795.
[17] 孙艳发. 基于全基因组关联研究技术筛选鸡产肉和肉品质性状相关候选基因[D]. 扬州: 扬州大学, 2013.
SUN Y F. Filtration of candidate gene related to meat production and quality traits based on genome-wide association study technique in chickens[D]. Yangzhou: Yangzhou University, 2013. (in Chinese)
[18] DALGAARD P. R Development Core Team (2010): R: a language and environment for statistical computing. 2010.
[19] NICODEMUS K K, LIU W, CHASE G A, TSAI YY , FALLIN M D. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genetics, 2005, 6(Supplement 1):S78. doi: 10.1186/1471-2156-6-S1-S78 .
[20] PRICE A L, PATTERSON N J, PLENGE R M, WEINBLATT M E, SHADICK N A, REICH D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 2006, 38(8): 904-909. doi:10.1038/ng1847.
[21] SUN Y, ZHAO G, LIU R, ZHENG M, HU Y, WU D, ZHANG L, LI P, WEN J. The identification of 14 new genes for meat quality traits in chicken using a genome-wide association study. BMC Genomics, 2013, 14: 458. doi:10.1186/1471-2164-14-458.
[22] ZHU F, CHENG S R, YANG Y Z, HAO J P, YANG F X, HOU Z C. Genome-wide association study of growth and feeding traits in Pekin ducks. Frontiers in Genetics, 2019, 10: 702. doi:10.3389/fgene.2019. 00702.
[23] DENG M T, ZHU F, YANG Y Z, YANG F X, HAO J P, CHEN S R, HOU Z C. Genome-wide association study reveals novel loci associated with body size and carcass yields in Pekin ducks. BMC Genomics, 2019, 20(1): 1. doi:10.1186/s12864-018-5379-1.
[24] DENG M T, ZHANG F, ZHU F, YANG Y Z, YANG F X, HAO J P, HOU Z C. Genome-wide association study reveals novel loci associated with fat-deposition and meat-quality traits in Pekin ducks. Animal Genetics, 2020, 51(6): 953-957. doi:10.1111/age.12995.
[25] LI G S, LIU W W, ZHANG F, ZHU F, YANG F X, HAO J P, HOU Z C. Genome-wide association study of bone quality and feed efficiency-related traits in Pekin ducks. Genomics, 2020, 112(6): 5021-5028. doi:10.1016/j.ygeno.2020.09.023.
[26] ZHU F, CUI Q Q, YANG Y Z, HAO J P, YANG F X, HOU Z C. Genome-wide association study of the level of blood components in Pekin ducks. Genomics, 2020, 112(1): 379-387. doi:10.1016/j.ygeno. 2019.02.017.
[27] LI G S, ZHU F, ZHANG F, YANG F X, HAO J P, HOU Z C. Genome-wide association study reveals novel loci associated with feeding behavior in Pekin ducks. BMC Genomics, 2021, 22(1): 334. doi:10.1186/s12864-021-07668-1.
[28] LIU D P, FAN W L, XU Y X, YU S M, LIU W J, GUO Z B, HUANG W, ZHOU Z K, HOU S S. Genome-wide association studies demonstrate that TASP1 contributes to increased muscle fiber diameter. Heredity, 2021, 126(6): 991-999. doi:10.1038/s41437-021- 00425-w.
[29] WANG L C, RUAN Z T, WU Z W, YU Q L, CHEN F, ZHANG X F, ZHANG F M, LINHARDT R J, LIU Z G. Geometrical characteristics of eggs from 3 poultry species. Poultry Science, 2021, 100(3): 100965. doi:10.1016/j.psj.2020.12.062.
[30] STODDARD M C, YONG E H, AKKAYNAK D, SHEARD C, TOBIAS J A, MAHADEVAN L. Avian egg shape: Form, function, and evolution. Science, 2017, 356(6344): 1249-1254. doi:10.1126/ science.aaj1945.
[31] LIN R L, CHEN H P, ROUVIER R, MARIE-ETANCELIN C. Genetic parameters of body weight, egg production, and shell quality traits in the Shan Ma laying duck (). Poultry Science, 2016, 95(11): 2514-2519. doi:10.3382/ps/pew222.
[32] DUMAN M, ŞEKEROĞLU A, YıLDıRıM A, ELEROĞLU H, CAMCı. Relation Between Egg Shape Index and Egg Quality Characteristics. Stuttgart: Verlag Eugen Ulmer, 2016. doi:10.1399/ eps.2016.117.
[33] RIZZI C. Yield performance, laying behaviour traits and egg quality of purebred and hybrid hens reared under outdoor conditions. Animals, 2020, 10(4): E584. doi:10.3390/ani10040584.
[34] WANG X, WANG H, ZHANG R, LI D, GAO M Q. LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells. International Journal of Biological Sciences, 2020, 16(2): 251-263. doi:10.7150/ijbs.38214.
[35] CHEN J, LAN J, YE Z, DUAN S, HU Y, ZOU Y, ZHOU J. Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma. Experimental Biology and Medicine (Maywood, N J), 2019, 244(14): 1137-1143. doi:10.1177/ 1535370219874339.
[36] LI S J, WU D, JIA H Y, ZHANG Z R. Long non-coding RNA LRRC75A-AS1 facilitates triple negative breast cancer cell proliferation and invasion via functioning as a ceRNA to modulate BAALC. Cell Death & Disease, 2020, 11: 643. doi:10.1038/s41419- 020-02821-2.
[37] BERTONI A P S, BRACCO P A, DE CAMPOS R P, LUTZ B S, ASSIS-BRASIL B M, DE SOUZA MEYER E L, SAFFI J, BRAGANHOL E, FURLANETTO T W, WINK M R. Activity of ecto-5'-nucleotidase (NT5E/CD73) is increased in papillary thyroid carcinoma and its expression is associated with metastatic lymph nodes. Molecular and Cellular Endocrinology, 2019, 479: 54-60. doi:10.1016/j.mce.2018.08.013.
[38] SUN W, YAO L, JIANG B, SHAO H, ZHAO Y, WANG Q. A role for Cdkl1 in the development of gastric cancer. Acta Oncologica (Stockholm, Sweden), 2012, 51(6): 790-796. doi:10.3109/0284186x. 2012.665611.
[39] LIU H, WANG L, GUO Z, XU Q, FAN W, XU Y, HU J, ZHANG Y, TANG J, XIE M, ZHOU Z, HOU S. Genome-wide association and selective sweep analyses reveal genetic loci for FCR of egg production traits in ducks. Genetics, Selection, Evolution, 2021, 53(1): 98. doi:10.1186/s12711-021-00684-5.
[40] 蒋晶晶. 三种家禽蛋壳厚度整齐性及蛋壳形状指标的研究[D]. 杭州: 浙江农林大学, 2020.
JIANG J J. The uniformity of eggshell thickness and eggshell shape indicators of three poultry[D]. Hangzhou: Zhejiang A & F University, 2020. (in Chinese)
[41] ZHANG Y N, DENG Y Z, JIN Y Y, WANG S, HUANG X B, LI K C, XIA W G, RUAN D, WANG S L, CHEN W, ZHENG C T. Age-related changes in eggshell physical properties, ultrastructure, calcium metabolism-related serum indices, and gene expression in eggshell gland during eggshell formation in commercial laying ducks. Poultry Science, 2022, 101(2): 101573. doi:10.1016/j.psj.2021.101573.
[42] ZHANG F, YIN Z T, ZHANG J F, ZHU F, HINCKE M, YANG N, HOU Z C. Integrating transcriptome, proteome and QTL data to discover functionally important genes for duck eggshell and albumen formation. Genomics, 2020, 112(5): 3687-3695. doi:10.1016/j.ygeno. 2020.04.015.
[43] FORREST M P, HILL M J, QUANTOCK A J, MARTIN-RENDON E, BLAKE D J. The emerging roles of TCF4in disease and development. Trends in Molecular Medicine, 2014, 20(6): 322-331. doi:10.1016/ j.molmed.2014.01.010.
[44] ISMAIL A B, NAJI M ' S, NEBIH İ, TUNCEL G, OZBAKIR B, TEMEL S G, TULAY P, MOCAN G, ERGOREN M C. The expression profile of WNT/β-catanin signalling genes in human oocytes obtained from polycystic ovarian syndrome (PCOS) patients. Zygote (Cambridge, England), 2022, 30(4): 536-542. doi:10.1017/ s0967199422000028.
[45] CHUNG J, WANG X L, MARUYAMA T, MA Y Y, ZHANG X L, MEZ J, SHERVA R, TAKEYAMA H, LUNETTA K L, FARRER L A, JUN G R. Genome-wide association study of Alzheimer's disease endophenotypes at prediagnosis stages. Alzheimer's & Dementia, 2018, 14(5): 623-633. doi:10.1016/j.jalz.2017.11.006.
[46] IMBRICI P, NEMATIAN-ARDESTANI E, HASAN S, PESSIA M, TUCKER S J, D’ADAMO M C. Altered functional properties of a missense variant in the TRESK K^+ channel (KCNK18) associated with migraine and intellectual disability. Pflügers Archiv - European Journal of Physiology, 2020, 472(7): 923-930. doi:10.1007/s00424- 020-02382-5.
[47] CEJUDO-MARTIN P, YUEN A, VLAHOVICH N, LOCK P, COURTNEIDGE S A, DÍAZ B. Genetic disruption of the sh3pxd2a gene reveals an essential role in mouse development and the existence of a novel isoform of tks5. PLoS ONE, 2014, 9(9): e107674. doi:10. 1371/journal.pone.0107674.
[48] ELBITAR S, RENARD M, ARNAUD P, HANNA N, JACOB M P, GUO D C, TSUTSUI K, GROSS M S, KESSLER K, TOSOLINI L, DATTILO V, DUPONT S, JONQUET J, LANGEOIS M, BENARROCH L, AUBART M, GHALEB Y, ABOU KHALIL Y, VARRET M, EL KHOURY P, HO-TIN-NOÉ B, ALEMBIK Y, GAERTNER S, ISIDOR B, GOUYA L, MILLERON O, SEKIGUCHI K, MILEWICZ D, DE BACKER J, LE GOFF C, MICHEL J B, JONDEAU G, SAKAI L Y, BOILEAU C, ABIFADEL M. Pathogenic variants in THSD4, encoding the ADAMTS-like 6 protein, predispose to inherited thoracic aortic aneurysm. Genetics in Medicine, 2021, 23(1): 111-122. doi:10. 1038/s41436-020-00947-4.
[49] KARUNAJEEWA H, HUGHES R J, MCDONALD M W,SHENSTONE F S. A review of factors influencing pigmentation of egg yolks. World's Poultry Science Journal, 1984, 40(1): 52-65. doi:10.1079/WPS19840006.
[50] COPPS K D, WHITE M F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia, 2012, 55(10): 2565-2582. doi:10.1007/ s00125-012-2644-8.
[51] SCHNEIDER A, ZHI X, MOREIRA F, LUCIA T, MONDADORI R G, MASTERNAK M M. Primordial follicle activation in the ovary of Ames dwarf mice. Journal of Ovarian Research, 2014, 7: 120. doi:10.1186/s13048-014-0120-4.
[52] THANGAVELU M, GODLA U R, PAUL S F D, MADDALY R. Single-nucleotide polymorphism of INS, INSR, IRS1, IRS2, PPAR-G and CAPN10 genes in the pathogenesis of polycystic ovary syndrome. Journal of Genetics, 2017, 96(1): 87-96. doi:10.1007/s12041-017- 0749-z.
[53] KUTTAPITIYA A, ASSI L, LAING K, HING C, MITCHELL P, WHITLEY G, HARRISON A, HOWE F A, EJINDU V, HERON C, SOFAT N. Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation. Annals of the Rheumatic Diseases, 2017, 76(10): 1764-1773. doi:10.1136/annrheumdis- 2017-211396.
[54] BRÉGEON M, TOMAS D, BERNAY B, ZATYLNY-GAUDIN C, GEORGEAULT S, LABAS V, RÉHAULT-GODBERT S, GUYOT N. Multifaceted roles of the egg perivitelline layer in avian reproduction: Functional insights from the proteomes of chicken egg inner and outer sublayers. Journal of Proteomics, 2022, 258: 104489. doi:10.1016/j. jprot.2022.104489.
[55] LESKO S L, ROUHANA L. Dynein assembly factor with WD repeat domains 1 (DAW1) is required for the function of motile cilia in the planarian. Development, Growth & Differentiation, 2020, 62(6): 423-437. doi:10.1111/dgd.12669.
[56] ELLINGHAUS E, ELLINGHAUS D, KRUSCHE P, GREINER A, SCHREIBER C, NIKOLAUS S, GIEGER C, STRAUCH K, LIEB W, ROSENSTIEL P, FRINGS N, FIEBIG A, SCHREIBER S, FRANKE A. Genome-wide association analysis for chronic venous disease identifies EFEMP1 and KCNH8 as susceptibility loci. Scientific Reports, 2017, 7: 45652. doi:10.1038/srep45652.
[57] SPENCER C C, SU Z, DONNELLY P, MARCHINI J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genetics, 2009, 5(5): e1000477. doi:10.1371/journal.pgen.1000477.
Genome-Wide Association Study of Egg Quality Traits in Longyan Shan-Ma Duck

1College of Life Sciences, Longyan University, Longyan 364012, Fujian;2Fujian Provincial Key Laboratory for the Prevention and Control of Animal Infectious Diseases and Biotechnology/Fujian Provincial Universities Key Laboratory of Preventive Veterinary Medicine and Biotechnology/Longyan University, Longyan 364012, Fujian;3Longyan Shan-Ma Duck Original Breeding Farm, Agricultural Bureau of Xinluo District, Longyan 364031, Fujian;4Fujian Provincial Animal Husbandry Headquarters, Fuzhou 350003;5College of Animal Science (College of Bee Science), Fujian Agriculture and Forestry University, Fuzhou 350002
【Objective】Single nucleotide polymorphisms (SNPs) and candidate genes for egg quality traits in duck were identified through genome-wide association study (GWAS) technology, so as to provide reference for molecular breeding to improve egg quality traits of Longyan Shan-Ma Duck.【Method】Egg quality traits of 235 female Longyan Shan-Ma Duck were measured, including egg weight (EW), egg shaped index (ESI), eggshell thickness (EST), eggshell strength (ESS), eggshell colour L* (ESCL), a* (ESCA), b* (ESCB), albumin height (AH), Haugh unit (HU), egg yolk colour (EYC), egg yolk weight (EYW), and egg yolk percentage relative to egg weight (EYP). Genetic parameters of these traits were estimated using multi-trait animal model by ASReml-R 4.1 software. Blood genomic DNAs of these ducks were genotyped using genotyping-by-sequencing (GBS) technology. The GWAS between egg quality traits of the late laying period and SNPs were performed. 【Result】 The heritability of EW, ESI, EST, ESL, ESA and AU was higher among the egg quality traits of Longyan Shan-Ma Duck, and ranged from 0.21 to 0.70. There was a strong positive genetic correlation (g= 0.91±0.37) between EW and AU, a strong negative genetic correlation (g= -0.98±1.03) between ESI and EYC. EST had a positive phenotypic correlation (p= 0.41±0.06) with ESS, negative genetic and phenotypic correlations with ESA (g= -0.86±0.25 andp= -0.15±0.07), and positive genetic and phenotypic correlations with ESB (g= 0.96±0.37 and 0.18±0.07). There were negative genetic and phenotypic correlations between ESA and ESB (g= -0.64±0.28 andp= -0.31±0.06). Results from the GWAS showed that seven SNPs were significantly associated with ESI, EST and yolk color (YC) at 5% Bonferroni-corrected genome-wide significance level (<4.74×10-6), involving six candidate genes. One SNP, chr20:11135563: G:C, was associated with ESI, which was in leucine rich repeat containing 75A gene, located on chromosome 20. Two SNPs, chr13:5766560:A:G and chrZ:968819:C:T, were associated with EST, which were located on chromosome 13, downstream 6.86 Kb ofand in transcription factor 4 gene, respectively. Four SNPs were associated with EYC, one SNP chr2: 38155965:G:A in potassium voltage-gated channel subfamily H member 8 gene located on chromosome 2; three SNPs located on chromosome 9, two SNPs, chr9:22623156:G:A and chr9:22623155:T:C, in insulin receptor substrate 1 gene, and one SNP, chr9:22490158:A:T, ingene. Eighty-one SNPs associated with egg quality traits reached at suggestive genome-wide significance level (<9.48×10-5) were also found. Thirteen SNPs associated with YC were distributed in the 0.84 Mb (22.16-23.00 Mb) region of chromosome 9.【Conclusion】In this study, genetic parameters of egg quality traits of Longyan Shan-Ma Duck were estimated. Seven significant SNPs, six candidate genes, and one candidate region affecting ESI, EST and EYC traits were identified through GWAS. The findings from the present study provided a reference for the molecular breeding of egg quality traits in Longyan Shan-Ma Duck.
Longyan Shan-Ma Duck; egg quality traits; genetic parameter; SNPs; GWAS

10.3864/j.issn.0578-1752.2023.03.014
2021-10-20;
2022-11-16
福建省种业创新与产业化工程(2021-2025)农业良种重大科研育种攻关与产业化工程项目(zycxny20211014)、福建省科技厅对外合作项目(2021I0045)、福建省科技厅引导性科技项目(2020N0034)、龙岩学院科研博士启动基金(LB2019001)
孙艳发,Tel:18250071633;E-mail:boysun2010@163.com。通信作者江宵兵,Tel:13960752743;E-mail:fzjxb@163.com。通信作者李焰,Tel:13860217279;E-mail:529783204@qq.com
(责任编辑 林鉴非)
