Similarity of Binding Potentials Between Plant DUF538 and Animal Lipocalin: Cholesterol Binding Ability of DUF538*
2023-02-26AshrafGHOLIZADEH
Ashraf GHOLIZADEH
(Division of Biochemistry,Department of Animal Sciences,Faculty of Natural Sciences,University of Tabriz,Tabriz,Iran)
Abstract Objective DUF538 (domain of unknown function 538) domain containing proteins are known as putative hypothetical proteins in plants.Until yet,there is no much information regarding their structure and function.Methods In the present research work,the homologous structures and binding potentials were identified between plant/mammalian lipocalins and plant DUF538 protein by using bioinformatics and experimental tools including molecular dynamics simulation,molecular docking and recombinant technology-based techniques.Results Molecular docking analysis of their interactions with lipidic ligands including cholesterol and palmitic acid revealed the similar and comparable binding potentials between DUF538 and lipocalin proteins.Both the test proteins were found to have more affinity to cholesterol molecule in compare to palmitic acid.By using recombinant technology-based experiments,the heterologously expressed and purified fused product of DUF538 protein exhibited about 61% cholesterol binding ability.Conclusion As a conclusion,plants DUF538 protein family was predicted to be the structural and may be the functional homologues of plants/animals lipocalin superfamily.
Key words binding potential,cholesterol,DUF538,lipocalin,palmitic acid
DUF538 (domain of unknown function 538) domain containing proteins belong to a large group of uncharacterized protein families as putative hypothetical proteins in plants.They have been found in more than 40 plant species,almost exclusively inEmbryophytaincluding the wide ranges of monocotyledonous and dicotyledonous plants,with poorly understood function[1-3].Their molecular mass is about 19-21 ku encoding around 170 amino acids.The three-dimensional structures of DUF538 proteins are dominated by β-strands (PDB ID: D1ydua1).DUF538 domain containing proteins or transcripts have often been identified from plants challenged with various abiotic and biotic environmental stresses including drought,elevated temperatures,nutrient deficiency,mixed elicitors,fungal pathogens,nematodes andMeloidogyneinfection[1,3-4].In general,DUF families,encompassing more than 22% of the entire Pfam database,have no yet clarified function.But based on the recent studies,they might play important roles in different plant stress responses[3,5-7].The involvement of DUF538 proteins in plant growth and developments has also been recently investigated,highlighting the importance of DUF538 protein family in higher plants[8].
In a different structural and functional studies by our research team,DUF538 proteins have been suggested to be the structural homologues of BPI (bactericidal/permeability increasing) proteins in the innate immune system of mammalians[9].Their binding potentials to the lipid A moiety of LPS (lipopolysaccharides) on the outer leaflet of the bacterial membranes (as like as BPI of immune system) and lipidic chlorophyll molecules in plants has been later on proposed[10-11].Very recently,the new suggestion of binding capacity of DUF538 proteins towards methylester compounds such as pectin molecules has been reported[12].These information led to the conduction of the present research to investigate more about the binding properties of DUF538 domain containing proteins with regards to the lipid/lipid like molecules.By using bioinformatics tools,we found that DUF538 proteins are structurally similar to lipocalins (lipid binding anticalin-like protein family).
Lipocalins are a large and ever expanding group of proteins that exhibit variable structural and functional properties,including lipid binding capacity.The physiological function of lipocalins is usually lies in the transport or storage of hydrophobic compounds such as retinol,vitamins,lipids and steroids[13-16].Based on subcellular distribution data analysis,human lipocalins may transfer signals from the extracellular space to the nucleus[17].They may also be as biomarkers or metabolic modulators[16,18].Lipocalins are mostly secreted proteins and have been characterized from variable prokaryotic and eukaryotic organisms including plant species[15,19-20].Their molecular mass is about 20 ku and their length ranges from 160 to 180 amino acids.The amino acid sequence identity between different lipocalin homologues,with pair wise comparisons,is often falling below 20%.In contrast with their low conservation at the sequence level,they have got conserved folding patterns/three-dimensional structures including eight-stranded anti parallel β-barrel,a short 310helix and an α -helix at their N-terminus[21].This β-dominated structures with flattened or elliptical shape usually encloses an internal ligand-binding site in lipocalin members.The lipocalins are best known for their binding of a remarkable array of small endogenous and exogenous hydrophobic ligands.A large cup-shaped cavity within the β-barrel and a loop scaffold at its entrance are well adapted for ligand binding.To accommodate ligands of different size and shape,the binding sites of different lipocalins are quite varied[21-22].Besides ligand binding ability,based on experimental evidences,lipocalins have been shown to bind to specific cell-surface receptors and internalized by receptor-mediated endocytosis[21,23].In plants,the first evidence for the presence of the putative lipocalin was reported by Bugoset al.[24].Then the first true plants lipocalins were identified from wheat (Triticum aestivum) andArabidopsis(Arabidopsis thaliana)[19].Finally,using the approaches of data mining of expressed sequence tag (EST) databases,other plant lipocalins with putative functions and cellular locations were identified and characterized[19].
Considering these together,to increase our knowledge about the structure and function of DUF538 proteins,our attempt was made to identify the similarity of binding potentials of DUF538 and lipocalin proteins with regard to cholesterol and palmitic acid molecules.Using computational and recombinant-based experimental approaches,the lipid-binding ability of DUF538 proteins was investigated and contributed to the novel research challenges of DUF538 protein family in plants.It was speculated that similar to lipocalins,DUF538 proteins may also bind to cholesterol molecules to contribute its trafficking mechanism or metabolizing in plants.
1 Materials and methods
1.1 Computational materials and analysis
The protein sequences ofArabidopsisDUF538 (Acc.no.AT5G01610),Arabidopsislipocalin (Acc.no.NP_200615) andGorilla gorillalipocalin (Acc.no.XP_004048725) were extracted from protein database,NCBI (national center for biotechnology information).The pair wise comparison of protein sequences were carried out by CLASTALW server at http://www.genome.ad.jp.Prediction of proteins tertiary structures were performed by Phyre2 internet-based software.For structural stability analysis,proteins were used as input for molecular dynamics simulation performed using GROMACS 2021.5 software.Input structures were prepared with Amberff99SB+ILDN force field.The correct position of hydrogen for all charged amino acids was determined using PROPKA software and the charge of the system was neutralized by adding chlorine ions.The protein was placed in a cubic box using gmx editconf software.The distance between the protein and the walls of the box was considered to be at least 1 nm.Next,TIP3P water molecules were added to the box using the gmx solvate.Energy minimization was performed on the structures using the steepest descent method in order to eliminate steric clashes and inappropriate geometry.In the next step,the system reached equilibrium in NVT (constant temperature and volume) conditions at 298 K in a period of 200 ps in a constant volume using the V-rescale algorithm,and then in NPT conditions at a constant pressure of 1 bar in a period of 200 ps using Parrinello-Rahman algorithm.Finally,the production stage was completed in NPT (constant temperature and pressure) conditions and within 100 ns.van der Waals interactions were calculated with a cut-off equal to 10 Å.Long-range electrostatic interactions were calculated by PME (Particle Mesh Ewald) method.LINCS algorithm was also used to constrain covalent bonds between heavy atoms and hydrogen.All simulations were done in periodic boundary conditions.Finally,the simulation information was stored in 4 ps intervals for analysis.Protein motif analysis was performed by using online motif search tool at http://www.genome.jp.
The interaction of the ligands,including cholesterol and palmitic acid,with the target proteins were performed by molecular docking to determine the binding energy and binding sites of the ligands on test proteins.AutoDock version 4.2 software was used for molecular docking performance.PDB for protein and ligand structures were obtained from Rcsb and PubChem databases,respectively.The structures of the test proteins were used as AutoDock input after modifications including heteroatom removal and modeling of deleted protein regions.After opening the structures with AutoDock software,first the water molecules were selected and removed.Then all the hydrogen atoms were added to the structures and finally all these changes were stored on the structures.Next,a grid boxes were created to select the docking locations on the proteins.For this,the entire protein was placed inside a box.At final step,the proteins as well as the ligand molecules are selected and the Lamarkian Genetic Algorithm (LGA),which determines the number of structural scans,was configured for use at this stage.Docking parameters were also created and the AutoDock program was run and saved all the output conformations as dlg file.After docking,the dlg file was opened and analyzed using AutoDockTools software.The conformations inside this file were ranked based on their binding energy.By using clustering system the binding sites were analyzed.
1.2 Experimental materials
Arabidopsis thalianaplants were prepared from Genetic Engineering Laboratory,Department of Plant Breeding and Biotechnology.Total RNA extraction from the leaves of test plant was carried out by Fermentas Trizol reagent (Cat.no.RN7713C;RNXTM).mRNA purification was done by using QIAGEN mRNA mini preparation kit (Cat.no.70022).Promega AcessQuickTMRT-PCR System was used for reverse-transcription PCR reaction (Cat.no.A1701).pGEM-T easy plasmid vector for cloning of RT-PCR product was from Promega.Fermentas DNA extraction Kit (Cat.no.K0513) was used for the purification of DNA from the agarose gel.The New England protein fusion and purification system kit (Cat.no.E8000S) utilized for bacterial transformationbyE.colistrain TB1 and recombinant protein expression by pMALc2X plasmid vector.Materials and columns for the fusion protein isolation and purification and antibody against MBP (maltosebinding protein) were provided in protein fusion and purification system kit.Cholesterol with more than 99% grade was from Sigma (Cat.no.57-88-5).All the other chemicals used in this research project were of molecular biology grades.
1.3 Cloning and expression of DUF538 as MBPfused protein
The cloning of DUF538 cDNA fromArabidopsis thalianaleaves was carried out using the following primer pair.Forward: 5'actcgaattcgatcagatcttcaacaaggttgg3' and Reverse: 5'tctaggatcctcgactcggagaccatttct 3'.In order to do directional cloning in expression vector,EcoRI andBamHI restriction sites were designed and included at 5' ends of both primers.For RT-PCR based cloning,total RNA was isolated from 0.2 g of leaf material by homogenizing in 2 ml of Trizol reagent.RNA collection was carried out after addition of 200 μl of chloroform and an equal volume of isopropanol,respectively.mRNA population was purified from total RNA sample using oligo dTcolumns.According to kit protocol,the RT-PCR reaction was performed with 0.5 μg of mRNA sample in 25 μl of master mix and 1 μl of primer set incubated at 45℃ for 45 min and proceeded with PCR cycling.PCR was programmed with 25 cycles as follows: pre-denaturation stage at 95℃ for 3 min,(denaturation at 95℃ for 1 min,annealing at 56℃ for 1.5 min,extension at 72℃ for 2 min),and ended with 10 min final extension at 72℃.The amplified products were then extracted from the agarose gel and cloned in pGEM-T easy cloning vector.The cloned fragments proceeded for the partial DNA sequencing in Microsynth center,Switzerland.
For the expression of DUF538 cDNA as MBPfused product,theE.colifusion protein expression system was used as described in our previous work[1].Briefly,the RT-PCR amplification product,after digestion withEcoRI andBamHI restriction enzymes,ligated into the pMALc2X expression vector and transferred intoE.coliTB1 competent cells.The transformed cells were plated on LB-agar medium (supplemented with ampicillin and X-gal) at 37℃ and a white recombinant clone was selected for the next protein extraction and purification processes.
1.4 Fused protein extraction and purification
In order to extract the fused product from recombinant bacteria,after growing the cells in rich broth medium containing glucose and ampicillin,the expression of fused product was induced by the addition of IPTG at 0.3 mmol/L final concentration for 8 h.Then,the expressing cells after precipitation by centrifugation at 4 000×gfor 10 min,dissolved in 25 ml of extraction buffer consisting of 20 mmol/L of Tris-Cl,200 mmol/L of NaCl,1 mmol/L of EDTA,1 mmol/L of sodium azide,and 10 mmol/L of BME (beta mercaptoethanol).After freezing the cells in the same extraction buffer,they were sonicated in short pulses of 15 s and centrifuged at 10 000×gat 4℃ for 20 min.The supernatant was utilized as crude protein extract of fused DUF538 protein.The purification process of fused protein was carried out by using the single-step maltose-binding affinity chromatography.For this purpose,the chromatography column was packed with amylase resin specific for the maltosebinding protein.The bound MBP-fused protein was eluted out from the amylase column by using the specific elution buffer containing crude protein extraction buffer plus 10 mmol/L maltose.The eluted product was analyzed by separating on 10% polyacrylamide gel using SDS-PAGE[25].
1.5 Western blotting
By using anti-body against maltose-binding protein,the Western analysis was carried out on the induced protein extract.Briefly,the total protein extract was separated on 10% SDS-PAGE and then transferred onto nitrocellulose membrane using blotting buffer containing 0.025 mol/L Tris-Cl pH 8.3,0.192 mol/L glycine and 20% ethanol.The blot was then kept in TBS-BSA buffer (0.02 mol/L Tris-Cl,pH 7.5,0.5 mol/L NaCl,1% BSA) at 4℃ and incubated with anti-MAL (maltodextrin-binding protein) antiserum for 2 h at 1∶500 ratio.After washing with TBS-T (TBS+0.05% Tween 20),the membrane was incubated with alkaline phosphatase conjugated goat-anti rabbit antibody (GARXAP;diluted 1:20 000 in TBS-BSA buffer) for 1 h.The signal band was visualized using a substrate solution containing 0.33 g/L NBT (nitro blue tetrazolium) and 0.165 g/L BCIP (5-bromo-4-chloro-3-indolyl phosphate) in 0.1 mol/L Tris-Cl buffer containing 0.1 mol/L NaCl and 5 mmol/L MgCl2.
1.6 Cholesterol binding ability test of DUF538 fused protein
To analyze the cholesterol binding ability of fused DUF538 protein,about 10 mg/L of purified fused product was dissolved in a buffer consisting of 20 mmol/L of Tris-Cl,200 mmol/L of NaCl,1 mmol/L of EDTA and then mixed with amount of cholesterol.The mixture was incubated under laboratory temperature for 20 min.The total cholesterol content of test sample was assessed by using cholesterol oxidase colorimetric method before and after mixing with fused protein and passing through maltose affinity column.For this,1 ml of cholesterol assay kit reagent was added to 10 μl of each test sample separately and incubated at 37℃ for 10 min to form the pink colored quinone complexes.The absorbance of the samples was then measured at 546 nm and the changes in the absorbance of test samples was estimated and presented as the approximate binding capacity of fused DUF538 towards cholesterol molecules.To test the possible role of MBP in cholesterol binding ability,another control sample including MBP-cholesterol mixture was considered.Each experiment was carried out with three replications.Data points represented on the graphs are the mean±SD (standard deviation) of replicates of each test.
2 Results
2.1 Structural similarity analysis
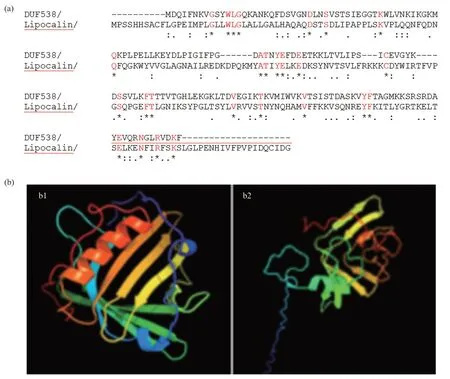
Fig.1 Amino acid sequence alignment and 3D comparison
In this study,the protein sequences ofArabidopsis thalianaDUF538 (AT5G01610),Arabidopsis thalianalipocalin (NP_200615) andGorilla gorillalipocalin (XP_004048725) were extracted from NCBI database.Their amino acid sequences were aligned by CLASTALW sequence comparing online server and the obtained results revealed that these proteins are not significantly identical in their primary structures.The homology scores between DUF538 and lipocalin proteins were predicted to be about 20%-22% (Figure 1a).However,the three-dimensional structural analysis by Phyre2 online server indicated that despite primary structures,the tertiary folds do much more support the presence of the highly homologous regions between DUF538 and lipocalin proteins.Comparison of the output data revealed that the folding patterns of these two proteins are confidently dominated by 7-8 anti parallel β-strands with the same organizations and orientations (Figure 1b).The presence of an alpha helical structure at the N-terminus ends is also identical between test proteins,highlighting their three-dimensional structural similarities.The stabilities of 3D structures of test proteins were analyzed by molecular dynamics simulation using GROMACS 2021.5.Analysis of the RMSD (root mean-square deviation) of α carbon atoms of both proteins revealed that lipocalin has lower constant value than that of DUF538 protein (0.22 nm versus 0.52 nm).In overall,data showed that both test proteins have stable structures during 100 ns simulation period.However,DUF538 structure is shown to be slightly fluctuated at starting time (Figure 2a).Analysis of secondary structural changes using DSSP software during molecular dynamics time revealed that lipocalin has low level of secondary structure fluctuation than DUF protein (one region versus two regions) and its structure is stable than DUF538 (Figure 2b).This analysis revealed that both test proteins share the similar pattern of β-plated structures.Comparison of structural conformations before and after molecular dynamics simulations revealed that lipocalin 3D structure is completely stable during simulation time (Figure 2c).But,the 20-25 residues of N-terminus part of DUF538 are structurally changed (Figure 2d).By a survey in protein motif databases using motif search tool at http://www.genome.jp,it was clarified that DUF538 and lipocalin proteins don not contain extra motif and their main motifs are DUF538 and lipocalin respectively (Figure 3).Thereby,it is speculated that these motifs may be the substitutes of each other.
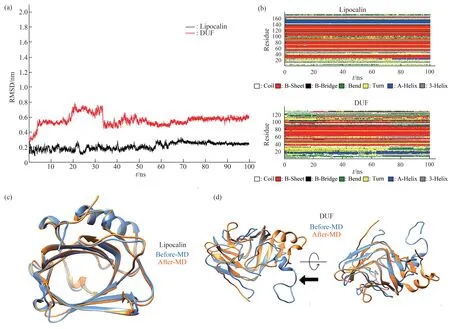
Fig.2 RMSD,secondary and 3D structural changes during molecular dynamics simulation
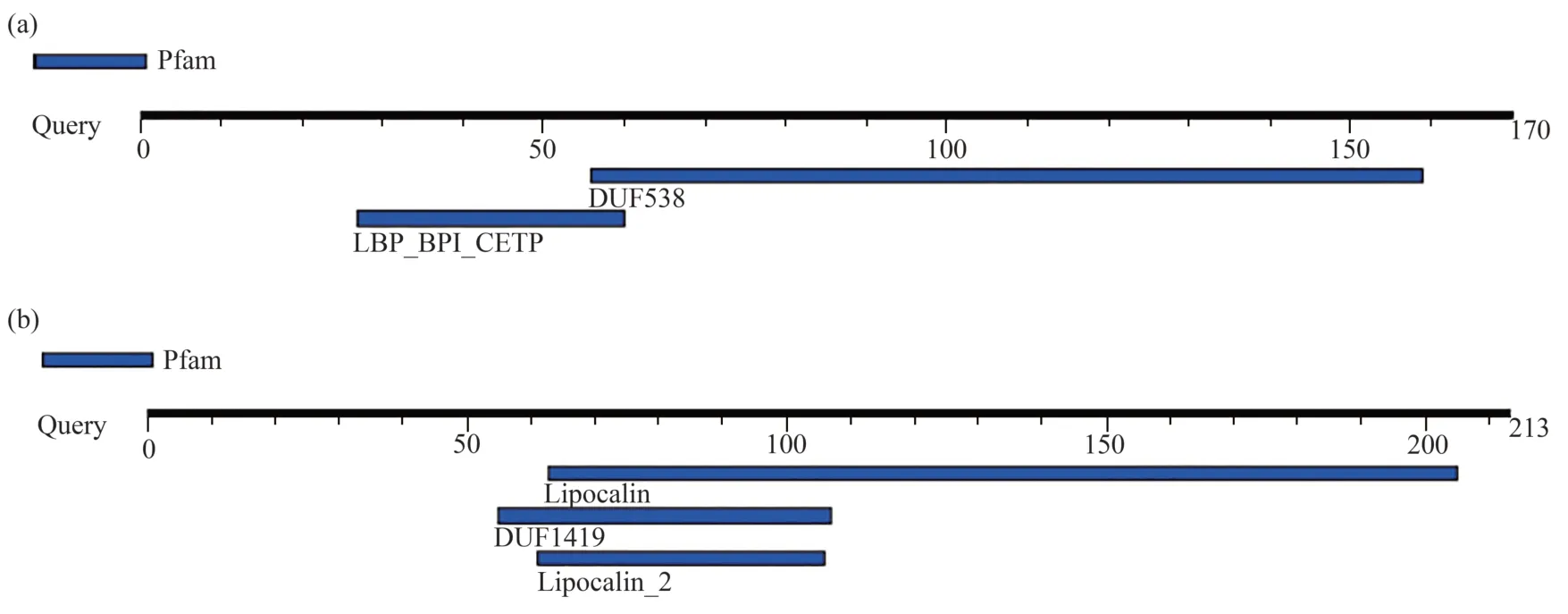
Fig.3 Protein motif analysis
2.2 Binding potentials similarity analysis
In this part of study,AutoDock4.2 and AutoDockTools were used to investigate and analyze the binding of test ligands (cholesterol and palmitic acid) to target proteins including DUF538 and lipocalin.For this,PDB for proteins and ligands structures were obtained from Rcsb and PubChem databases,respectively.Then,using AutoDockTools,polar hydrogens were added and Gasteiger charges were placed on all atoms and rotatable bonds were calculated.Grid box was selected with a distance of 0.375 Å and the target protein was considered inside the box with the dimensions of 126×126×126 and the number of 300 runs.Figure 4 shows the number of clusters,the number of structures in each cluster,and the amount of best binding energy for each cluster in DUF538-cholesterol and DUF538-palmitic acid interactive complexes.According to the obtained results,cluster 1 in the DUF538-cholesterol complex has 113 structures and the binding energy of -8.4 kcal/mol.In the diagram related to DUF538-palmitic acid complex,cluster 1 has 55 members with the binding energy of about -5.4 kcal/mol.Thereby,the results reveals that DUF538 protein has got more potential to bind to cholesterol molecule in compare to palmitic acid.The various energies including electrostatic and van der Waals energies as well as the inhibitory constants of cholesterol and palmitic acid-DUF538 complexes for the first three clusters are shown in Table 1.As compared to DUF-palmitate,DUF-cholesterol complexes show higher number of interactive structures (174 versus 98) and binding energy (24.11 kcal/mol versus 16 kcal/mol).Besides this,the difference between inhibitory constant values of DUF-cholesterol and DUF-palmitic complexes (4.202 μmol/L versus 370.11 μmol/L) is very significant and more considerable.In compare to van der Waals energy,the amount of electrostatic energy related to the binding of cholesterol to DUF538 protein is very low (0.18 versus 6.98),showing the absence of a charged functional group in the structure of the cholesterol molecule.According to the results,comparing the binding potentials (particularly including binding energies and inhibitory constant values) of DUF538 protein to cholesterol and palmitic acid indicates that DUF538 has more affinity to cholesterol molecule than palmitic acid.
In the next step,the binding sites of DUF538 protein to the test ligands were analyzed.Figure 4 shows the binding sites and the surrounding amino acids in two clusters related to the binding of cholesterol and palmitic acid molecules to DUF protein.Chimera and LigPlus softwares were used to prepare 3D and 2D image,respectively.As shown,both the test ligands (including cholesterol and palmitic acid) are mostly bound to the N-terminus part of DUF protein in different complexes.In particular,the results indicated that the conserved N-terminus loop and helical structure of DUF538 protein has decisive role in ligand-protein complex formation (Figure 4).Analysis of interactive amino acids showed that the amino acids Leu87,Pro89,Asn74,Asp71,Trp41,Met50,Lys45,Lys47,Ile46,Asp168,Ile88,Thr106 and Thr107 are involved in DUFcholesterol complex formation in cluster 1.In this cluster,Thr107 has been shown to interact with hydrogen bonds.In cluster 2,the amino acids Thr106,Thr107,Ile88,Leu87,Pro55,Met50,Asn74,Gly70,Gly48,Asp71,Asp168 and Pro89 are involved in complex formation.In case of DUF-palmitic acid complex,the amino acids Gly48,Lys47,Met50,Ile46,Asn44,Lys45 and Trp41 are involved in complex formation in cluster 1,in which the amino acids Gly48 and Lys47 are interacted with hydrogen bonds.In cluster 2,the amino acids Ser100,Lys126,Trp130,Thr125,Val101,Lys103,Glu93,Lys124 and Leu102 are involved.In this cluster,the amino acids Ser100 and Lys126 are interacted with hydrogen bonds.Analysis of data revealed that cholesterol and palmitic acid have shared the same bound amino acids (including Lys47,Met50,Ile46,Asn44,Lys45 and Trp41) in cluster 1.But,there are no conserved interactive amino acids between DUF complexes in cluster 2.Also data analysis revealed that the bound amino acids of DUF-complexes are not identical between two clusters.In general,analysis of the interactive amino acids in DUF-cholesterol and DUFpalmitic acid complexes showed that the N-terminus part of DUF538 protein is involved in the formation of both complexes and the primary structure of test protein has no much decisive role in ligand-protein interaction.
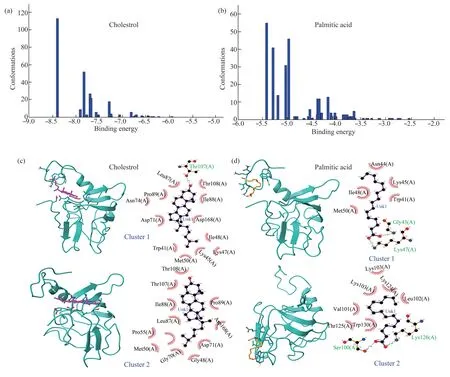
Fig.4 Binding energies and binding sites analysis of DUF538-ligand complexes
Similar to DUF538,the molecular docking results of lipocalin protein with cholesterol and palmitic acid molecules have also been investigated.According to the results,cluster 1 in lipocalincholesterol complex has 19 interactive structures and the binding energy of -8.3 kcal/mol.While,in the diagram related to lipocalin-palmitic acid complex,cluster 1 has 95 members with the binding energy of -6.99 kcal/mol (Figure 5).Thereby,the results reveal that such as DUF538,lipocalin protein has got more potential to bind to cholesterol molecule in compare to palmitic acid.The energy details and the binding potentials of the top 3 clusters obtained by molecular docking of cholesterol and palmitic acid with lipocalin protein are shown in Table 1.In compare to lipocalin-palmitic acid,the lipocalin-cholesterol complexes have got more binding energy (24.18 kcal/mol versus 20.24 kcal/mol) and low inhibitory constant value (3.961 μmol/L versus 31.11 μmol/L).Analysis of electrostatic energies values (0.15 versus 9.95) showed that in compare to van der Waals energies,electrostatic bonds have more decisive role in lipocalin-palmitate complex formation.The overall results revealed that similar to DUF538 protein,lipocalin has also more affinity to cholesterol molecule rather than palmitic acid.
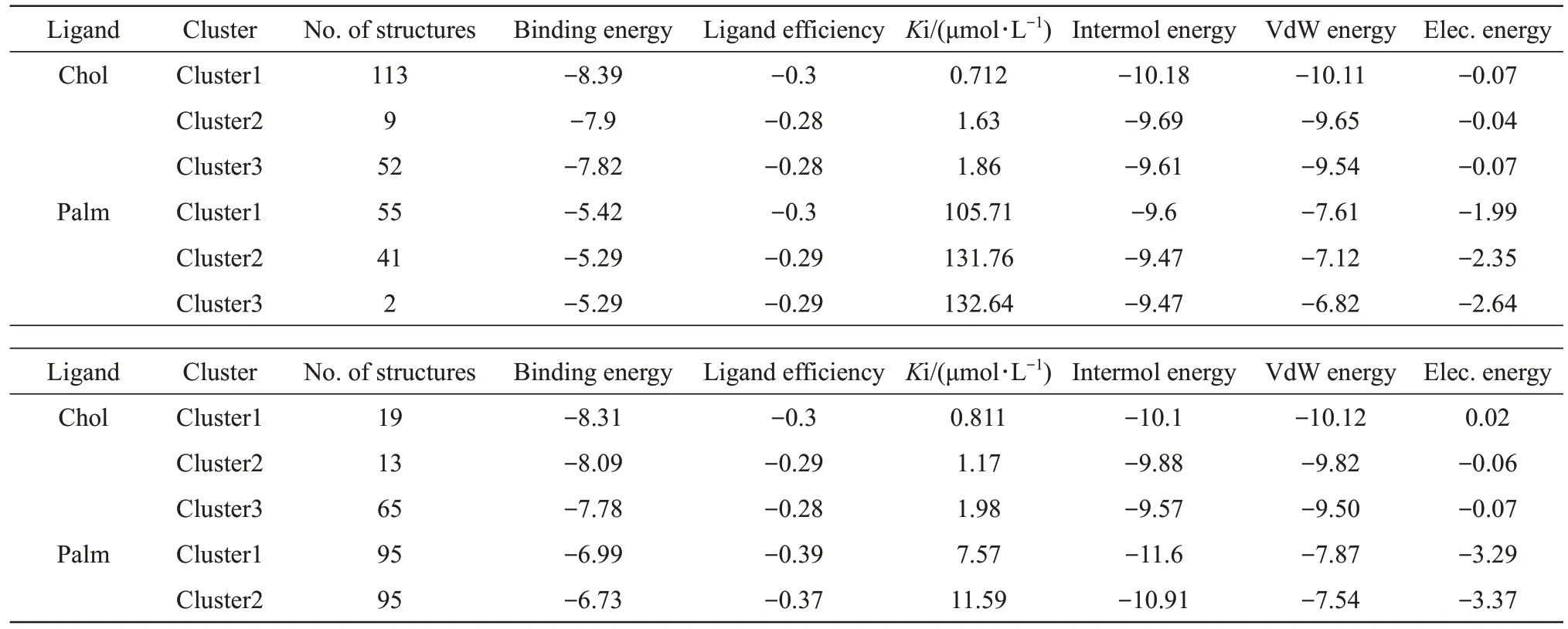
Table 1 Energy details of the top 3 clusters obtained by molecular docking of cholesterol and palmitic acid with DUF538 (upper) and lipocalin (lower) proteins
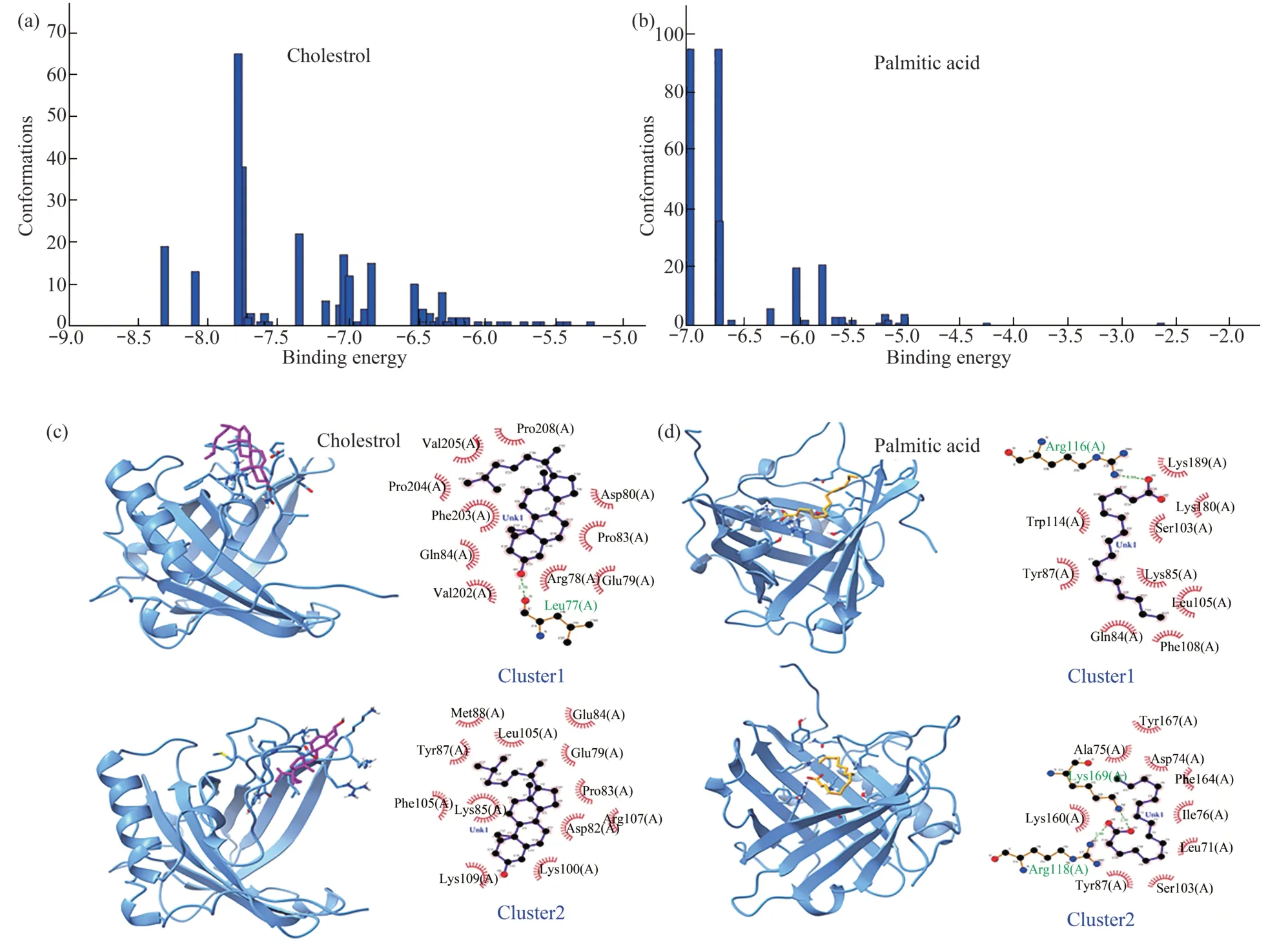
Fig.5 Binding energies and binding sites analysis of lipocalin-ligand complexes
Same to DUF538,the binding sites and the surrounding amino acids of cholesterol and palmitic acid-lipocalin complexes in clusters 1 and 2 were also investigated (Figure 5).In cluster 1 of lipocalincholesterol complex,the amino acids Leu77,Val202,Gln84,Phe203,Pro204,Val205,Pro206,Asp80,Pro83,Arg78 and Glu79 are involved.In cluster 2,the amino acids Met86,Leu105,Tyr87,Lys85,Phe105,Lys109,Lys100,Asp82,Arg107,Pro83,Glu79 and Glu84 constitute the binding amino acids.In case of lipocalin-palmitic acid complex formation,the amino acids Arg116,Trp114,Tyr87,Gln84,Phe106,Leu105,Lys85,Ser103,Lys160 and Lys169 are involved in cluster 1 in which Arg116 is interacted with hydrogen bonds.In cluster 2,the amino acids Arg116,Lys169,Lys160,Ala75,Tyr167,Asn74,Phe164,Ile76,Leu71,Ser103 and Tyr87 are involved in complex formation in which the amino acids Arg116 and Lys169 are interacted with hydrogen bonds.Data analysis showed that cholesterol and palmitic acid molecules bind to lipocalin protein at its interior parts,as compared to DUF538 protein complexes.Besides this,the comparison of interactive amino acids between four different lipocalin-ligand complexes indicated that the primary structure of test protein has no decisive role in ligand-protein interaction.
Finally,the comparison of DUF and lipocalin ligand-binding abilities indicated that both test proteins are able to bind to cholesterol and palmitic acid molecules with higher binding energy but no significant conserved binding sites.The primary and secondary binding sites were predicted to be varied between DUF lipocalin complexes.
2.3 Experimental analysis of DUF538 binding potential
For further confirmation of the docking results,the binding potential ofArabidopsisDUF538 protein towards cholesterol molecule was investigated by MBP-based recombinant technology.To do this,the cDNA of DUF538 was amplified fromArabidopsis thalianaleaves (grown under normal laboratory conditions) by RT-PCR using specific primer set as described under materials and methods section.The amplified product was analyzed on agarose gel.The results showed that the size and the partial sequence of the amplified cDNA were in perfect agreement with the expected size and the sequence of the already reportedArabidopsiscDNA (data not presented).The amplified cDNA was then restricted withEcoRI andBamHI enzymes (the restriction sites were already located on 5' ends of primers) and directionally ligated on pMALc2X expression vector and transformed into TB1 strain ofE.coli.The MBP-fused DUF538 protein was extracted and purified from TB1 expressing cells by using single-step maltose-affinity chromatography.To confirm the purity of the recombinant product,SDS-PAGE electrophoresis and Western blotting were performed using antibody against maltose-binding protein (Figure 6).The molecular size of the fused protein was analyzed to be about 62 ku on the gel.
In order to assess the cholesterol binding ability of MBP-fused product of DUF538,the free cholesterol content of test solution was assessed before and after mixing with purified fused DUF538 protein and passing through the maltose affinity column.The changes in the absorbance of test samples (before and after mixing with fused protein) was estimated and presented as the approximate binding capacity of fused DUF538 towards cholesterol molecules.The results showed that the absorbance of the test sample is decreased to 0.7 as compared to 1.8,the absorbance of the control samples.Accordingly,the amount of cholesterol was found to be sharply decreased to the level of 39% in comparison to the control (Figure 6).On the other hand,this data revealed that about 61% of the cholesterol molecules are absorbed by DUF538 protein.Comparison of the MBP containing control sample with MBP-free control and test sample showed that cholesterol binding ability is exerted from DUF538 moiety of the fused product and not from the MBP partner.On the other hand,MBP is not involved in cholesterol binding ability of DUF538.
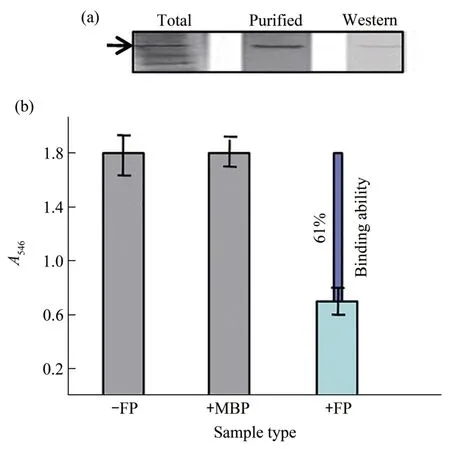
Fig.6 SDS-PAGE,Western analysis and cholesterol binding ability assessment
3 Discussion
The aim of this study as the first ever time research work was to analyze the structural similarity as well as the binding potentials between DUF538 and lipocalin proteins using computational as well as experimental tools including recombinant techniques.Based on our previous research results,DUF538 domain containing proteins were suggested to be able to bind to lipid-like molecules such as chlorophyll and lipopolysaccharides[10-11].Thereby,DUF538 protein family was proposed to be the structural/functional homologue of lipocalin (lipid binding anticalin like proteins) protein family in plants or animals.Our primary structural analysis did not support the significant structural similarity between DUF538 and plant/animal lipocalin proteins.In accordance with this,the previous studies on sequence and structure relationship been suggested that lipocalins display very low levels of sequence identities often falling below 20%[13].In contrast to their low level of conservation at the primary sequences,comparison of different lipocalins tertiary structures have shown that their overall folding pattern including 8 anti parallel β-stranded structure is highly conserved[17,21].Therefore,as a strong clue about the structural/functional similarities between selected test proteins,our searches were extended to identify and compare the tertiary structures of test proteins.The relevant computational analysis,about the similar β-dominated folding pattern,highlighted that DUF538 domain containing proteins might be the potential structural/functional homologues of lipocalins.Our motif analysis data also speculated that there are no extra main motifs on test proteins and DUF538 and lipocalin motifs may be the substitutes of each other.Lipocalin members are typically known as binding proteins.They have different molecular recognition properties including their binding to several lipid-like hydrophobic molecules (such as retinol,fatty acids and cholesterol),binding to specific cell-surface receptors and formation of different macromolecular complexes[22-23].Keeping this in view,for more similarity analysis,the binding potentials of DUF538 and lipocalin test proteins were investigated.The overall analysis of molecular docking results regarding the binding potentials of DUF538 and lipocalin proteins indicated that both these proteins are able to bind cholesterol and palmitic acid molecules.However,their abilities to interact with cholesterol molecule are more than palmitic acid.The N-terminus parts of both proteins were found to be the interactive sites for cholesterol and palmitic acid.All these data together revealed that DUF538 protein family can be considered as the potential structural and may be functional homologue of lipocalin superfamily.But to how extend this computational prediction can be reliable,was depended on experimental analysis,too.Therefore,following the computational clarifications we extended our analysis by recombinant-based experimental tool and examined the cholesterol binding ability of DUF538,in vitro.The experiment result confirmed the cholesterol binding capacity of DUF538 protein as it has already been reported for lipocalins[22].We selected MBP fusion technology since MBP fusion is known as the most efficient production and purification system of correctly folded proteins in heterologous cells,enhancing protein structural folding and stability in a native confirmation[26-28].Thereby,the biological activities of MBP-fused products including DUF538 are rarely influenced[1,29].The overall result obtained by this assay did more support our computational data indicating DUF538 family might be the potential functional homologue of lipocalin protein superfamily.
Our previous report suggested that plant DUF538 proteins are the structural/functional homologues of BPI (bactericidal/permeability increasing) proteins in the innate immune system of mammalians[9].They do affect the bacterial growth rates through the binding to the lipid A moiety of bacterial membranes lipopolysaccharides as like as BPI in mammalians immune system.On the other hand,the members of lipocalins are identified as innate immune proteins with bactericidal activity[30-31].This reveals that lipocalins and DUF538 are not extra domains,but they can be the potential substitute of each other that share the similar structure and function in different organisms including plants and animals.
The present work results indicated that similar to lipocalins,DUF538 domain containing proteins exhibit efficient binding capacity towards lipid-like cholesterol molecule.On the other hand,DUF538 proteins were previously found to be the functional homologues of WSCP (water soluble-chlorophyll binding proteins) and efficiently bind to chlorophyll molecules same to WSCP[11].Considering chlorophyll molecules containing lipid moiety in their structures,the variable lipid binding and transporting capacities is predicted for DUF538 as been already approved for lipocalins[22].Besides binding capacities,DUF538 and lipocalins are also proposed to exert hydrolytic activities towards bound molecules.DUF538 proteins have already been suggested to be structurally and functionally comparable to lipolytic hydrolases particularly esterase-lipases[10,12].This reveals that DUF538 protein family not only is able to bind lipid/lipid-like molecules but also it can use them as substrate for hydrolysis.The hydrolytic activity of DUF538 proteins towards chlorophyll molecules and methylester compounds have already been approved by our research team[10-12].Keeping this information in view and considering the structural similarities of DUF538 and lipocalin proteins,their hydrolytic capacities towards cholesterol molecules cannot be away from thinking.We hope the results of the present work will open the gate to specific and interesting studies on cholesterol transporting and metabolizing potencies of DUF538/lipocalin proteins in plant and animal systems.Binding proteins are essential for most cellular events and regulations.To exert their functions,they bind or interact with different other molecular partners that are important in understanding the mechanisms of action in biological world.Thereby,based on the binding abilities of plants DUF538 proteins,they can be nominated as novel binding proteins and may be used in plants,animal or medical biotechnology and drug development.
4 Conclusion
Considering the similar structure and binding potentials,plants DUF538 domain containing protein was proposed to be the structural and may be functional homologues of lipocalin proteins in plants and animals.They were suggested to be the potential substitute of each other and share the similar structure and function in different organisms.The identifications presented in this work will contribute to our future specific investigations and detailed understanding of the interactions of DUF538/ lipocalin proteins with cholesterol in plant and animal systems.
AcknowledgmentsThe author of this paper is thankful to the Department of Animal Sciences,Faculty of Natural Sciences,and Research Center for Bioscience and Biotechnology,University of Tabriz for the facilities and financial supports.