A multiplex real-time PCR assay for simultaneous detection of classical swine fever virus,African swine fever virus,and atypical porcine pestivirus
2023-02-03SONGXiangpengXlAYingjuXULuZHAOJunjieWANGZhenZHAOQizuLlUYebingZHANGQianyiWANGQin
SONG Xiang-peng,XlA Ying-ju,XU Lu,ZHAO Jun-jie,WANG Zhen,ZHAO Qi-zu,LlU Ye-bing,ZHANG Qian-yi,WANG Qin
China Institute of Veterinary Drug Control/National Reference Laboratory for Classical Swine Fever,Beijing 100081,P.R.China
Abstract With the implementation of the C-strain vaccine,classical swine fever (CSF) has been under control in China,which is currently in a chronic atypical epidemic situation.African swine fever (ASF) emerged in China in 2018 and spread quickly across the country.It is presently occurring sporadically due to the lack of commercial vaccines and farmers’ increased awareness of biosafety.Atypical porcine pestivirus (APPV) was first detected in Guangdong Province,China,in 2016,which mainly harms piglets and has a local epidemic situation in southern China.These three diseases have similar clinical symptoms in pig herds,which cause considerable losses to the pig industry.They are difficult to be distinguished only by clinical diagnosis.Therefore,developing an early and accurate simultaneous detection and differential diagnosis of the diseases induced by these viruses is essential.In this study,three pairs of specific primers and Taq-man probes were designed from highly conserved genomic regions of CSFV (5´ UTR),African swine fever virus (ASFV) (B646L),and APPV (5´ UTR),followed by the optimization of reaction conditions to establish a multiplex real-time PCR detection assay.The results showed that the method did not cross-react with other swine pathogens (porcine circovirus type 2 (PCV2),porcine reproductive and respiratory syndrome virus (PRRSV),foot-and-mouth disease virus (FMDV),pseudorabies virus (PRV),porcine parvovirus (PPV),and bovine viral diarrhea virus BVDV).The sensitivity results showed that CSFV,ASFV,and APPV could be detected as low as 1 copy mL-1;the repeatability results showed that the intra-assay and interassay coefficient of variation of ASFV,CSFV,and APPV was less than 1%.Twenty-two virus samples were detected by the multiplex real-time PCR,compared with national standard diagnostic and patented method assay for CSF (GB/T 27540-2011),ASF (GB/T 18648-2020),and APPV (CN108611442A),respectively.The sensitivity of this triple real-time PCR for CSFV,ASFV,and APPV was almost the same,and the compliance results were the same (100%).A total of 451 clinical samples were detected,and the results showed that the positive rates of CSFV,ASFV,and APPV were 0.22%(1/451),1.3% (6/451),and 0% (0/451),respectively.This assay provides a valuale tool for rapid detection and accurate diagnosis of CSFV,ASFV,and APPV.
Keywords: classical swine fever virus,African swine fever virus,atypical porcine pestivirus,real-time PCR
1.lntroduction
Classical swine fever (CSF) is caused by classical swine fever virus (CSFV),which is a severe and fatal infectious disease in pigs.With vaccines’ contribution to immunization and the implementation of prevention measures,CSF is under control,showing a chronic and atypical epidemic situation in some areas of China (Wang and Tu 2015;Tonget al.2020).African swine fever(ASF),caused by the African swine fever virus (ASFV),is an acute and severe infectious disease that can lead to high mortality in domestic pigs (Galindo and Alonso 2017;Brown and Bevins 2018).ASF was first detected in a pig farm in Shenbei District of Shenyang City,Liaoning Province,China,in August 2018;it then spread widely in China and has severely affected the pig industry development (Geet al.2018).CSF and ASF,animal diseases that are notifiable to the World Organization for Animal Health (OIE),are classified as one animal disease in China (Liet al.2017;Wuet al.2020).In 2015,researchers found a highly mutated RNA virus in American pig serum through metagenomic sequencing and named it Atypical porcine pestivirus (APPV) (Hauseet al.2015).APPV is considered to be the pathogen that causes congenital tremor (CT) type A-II in newborn piglets.The clinical symptoms of the disease are characterized by generalized body shaking with variable degrees of hypomyelination in brain and spinal cord (Shenet al.2018;Panet al.2019).Nowadays,CSF,ASF,and APPV are prevalent in many areas of China.They are detected in some small intensive pig farms,especially small and backyard pig farms in China,due to the weak awareness of prevention and common concurrent and secondary infections of two or more viral pathogens (Shiet al.2016;Ouyanget al.2019).CSF and ASF have similar clinical and pathological signs and thus are not easy to be distinguished clinically (Schulzet al.2017;Andraud and Rose 2020).In addition,CSF and APPV can both induce CT in newborn piglets,which is usually difficult to be identified through clinical manifestations (Dallet al.2020;Maliket al.2020).Therefore,differential diagnosis of the three diseases in laboratories is necessary.
The multiplex real-time PCR is characterized by high throughput detection capability and fast speed.Some researchers have developed multiplex PCR for detecting multiple pathogens.A previous study (Grauet al.2015)assessed the feasibility of simultaneous ASFV,CSFV,and FMDV detection by multiplex reverse transcription real-time PCR (mRT-qPCR) in swine oral fluids,which is critical to limit the impact and spread of these disease outbreaks.The ability to perform herd-level surveillance for three diseases rapidly and cost-effectively using a single diagnostic sample and test is highly desirable.Some studies established a multiplex real-time PCR quantitation of human herpesvirus-6,-7,-8 viruses:application in blood transfusions,which showed good specificity and sensitivity in blood transfusion screening(Zhenget al.2021).In order to monitor these diseases more effectively,it is vital to develop a fast,accurate,easy,and simple-to-handle method to differential detect them at the same time.This study focused the multiplex real-time PCR method that can simultaneously detect CSFV,ASFV,and APPV,and carried out preliminary evaluation and application.
2.Material and methods
2.1.Primers and probes
According to the conservative regions of CSFV,ASFV(Zsaket al.2005),and APPV genomes from GenBank(accession no.),the multiple pairs of primers and probes were designed respectively by the Primer 5 Software.After screening,the most suitable ones were selected,as shown in Table 1.Primers and probes were synthesized by Sangon Biotech (Shanghai) Co.,Ltd.,China.
2.2.Virus or clinical samples and nucleic acid extraction
This study prepared 22 samples,including CSFV (8),ASFV (3),APPV (2);other samples (PCV2,PRRSV,FMDV,PRV,PPV,BVDV) and negative samples (3)shown in Table 2 were used for the specificity and compliance test.Then,200 mL of each processed sample was used to extract nucleic acid using the Magnetic Viral DNA/RNA Kit (TIANGEN,China).Reverse transcription was performed using the PrimeScriptTMMaster Mix(TaKaRa Bio.,China) according to the manufacturer’sinstructions.The whole process was under aseptic operation to prevent sample contamination.DNA or cDNAs were then used as the multiplex real-time PCR template and temperately stored at -20°C.

Table 1 Primers and probes of CSFV,ASFV,and APPV
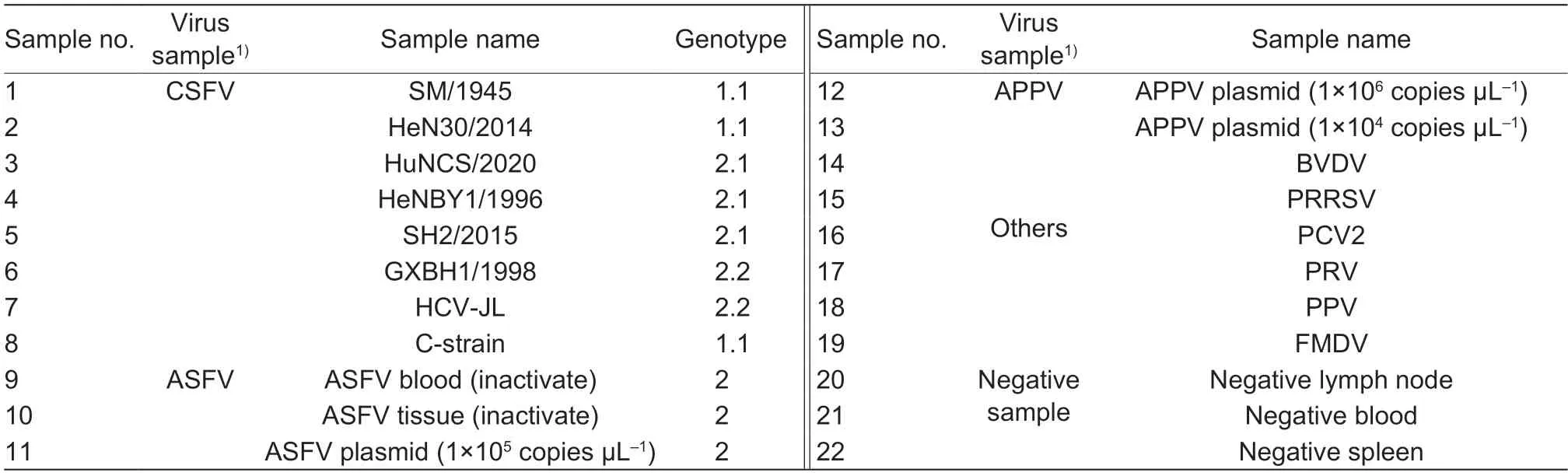
Table 2 Samples background information
2.3.Construction of recombinant plasmid
The genes of the conservative regions of CSFV (5´ UTR),ASFV (B646L),and APPV (5´ UTR) were cloned into a PUC57 vector by Sangon Biotech.In order to obtain a large number of positive plasmids,the constructed plasmids were subjected to positive transformation intoEscherichia coliDH5α competent cells (Sangon Biotech).Then,the bacterial solution was shaken and cultured for 12-16 h at 37°C to extract the plasmid using QIAprep®Spin Miniprep Kit (QIAGEN,Germany) and used a NanoDrop spectrophotometer (Thermo Fisher,USA) to quantify positive plasmids.Finally,singleplex real-time PCR experiments were performed to test the quality effect of positive plasmids.
The Dalton calculation method was used to calculate the copy number of positive plasmids,and the plasmid was diluted from 1×107copies mL-1to 1×10-1copies mL-1.The diluted plasmid was used as templates for singleplex and multiplex real-time optimization and sensitivity experiments of PCR conditions.The copy number of positive plasmids was calculated using the formula:
2.4.Optimization of the singleplex real-time PCR reaction conditions
The matrix method (Table 3) was used to optimize the annealing temperature,primer,and probe concentration.The total volume of the singleplex realtime PCR reaction is 25 μL,consisting of 12.5 μL 2×HyperProbe Mixture (Jiangsu CWBIO Co.,Ltd.,China),1 mL of positive plasmid template,and the sterilized nuclease-free water supplement to the final volume at the annealing temperatures (56,58,and 60°C).The following procedures were used to perform amplification on the LightCycler®480 II Instrument (Roche Life Sciences,Swiss): 95°C for 30 s;95°C for 10 s,annealing temperature and extension temperature (56,58,and 60°C) for 20 s,number of cycles for 45 cycles,collecting fluorescence;and 40°C for 30 s.The results were determined when the Ctvalue was 45 cycles to obtain the optimal reaction conditions for the singleplex real-time PCR.
2.5.Optimization of the multiple real-time PCR reaction conditions
Based on optimizing the singleplex real-time PCR reaction conditions for the multiple real-time PCR reactions,the matrix method (Table 4) was used again to optimize the concentration of primers and probes.Three different primers,probes,and templates were placed in one tube for a real-time PCR amplification.
2.6.Standard curves analysis
This study used 10-fold-diluted plasmids of each corresponding synthetic DNA from 1×107copies mL-1to 1×101copies mL-1(3 replicates for each gradient) for standard curve analysis.The amplification curve and standard curve were generated according to the optimized multiplex real-time PCR assay.The calculation ofR2(correlation coefficient) and standard equation was based on the amplification curve.
2.7.Specificity of the multiplex real-time PCR assay
Nineteen positive and three negative samples (shown in Table 2) were collected,and total nucleic acid (DNA/RNA) was extracted.The specificity of the established multiple real-time PCR was tested using DNA or cDNA of 22 samples to exclude potential false positives caused by other viruses that may exist in the sample.Referring to some literatures (Geet al.2018;Gilliauxet al.2019;Moliniet al.2020),we compared the availableB646Lgene sequences of ASFV in the NCBI GenBank and synthesized the plasmids of seven different genotypes ofB646Lfull-length sequences by Sangon Biotech.The genotypes include: genotype I,BA71V strain (GenBank no.NC_001659.2);genotype V,10 strains (GenBank no.AY578704.1);genotype VIII,SPEC_57 strain (GenBank no.MN394630.3);genotype IX,R7 strain (GenBank no.MH025917.1);genotype X,B53 strain (GenBank no.MT956648.1);genotype XXII,RSA_2008 strain (GenBank no.MN336500.3);genotype XXIII,ETH/3a strain(GenBank no.KT795357.1).We tested seven different ASFV plasmids with this method we developed.
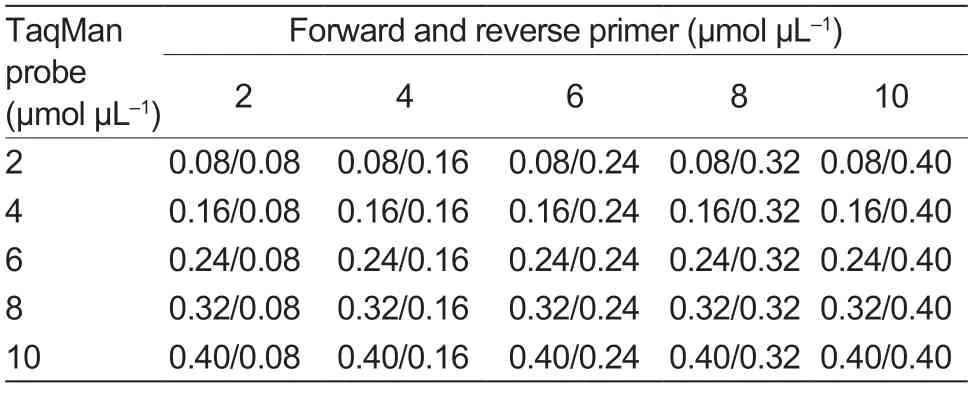
Table 3 Optimization of singleplex real-time PCR

Table 4 Optimization of multiple real-time PCR
2.8.Sensitivity of the multiplex real-time PCR assay
In order to assess its sensitivity and draw an amplification curve,three 10-fold-diluted positive plasmids from 1×105to 1×10-1copies mL-1(three replicates for each gradient)were used to amplify according to the optimized multiple real-time PCR reaction conditions.If there was a Ctvalue and a clear amplification curve,it was judged as positive;otherwise,it was a negative result.
2.9.Repeatability of the multiplex real-time PCR assay
In order to evaluate the repeatability of the assay,interand intra-assay tests were performed,three positive plasmids with different gradients were mixed together with gradients of 1×107,1×105,and 1×103copies mL-1as templates and then amplified by the established multiplex real-time PCR method.
2.10.Comparison of the multiplex real-time PCR with other detection methods
In order to compare the compliance of different detection methods,we used DNA or cDNAs of 22 samples (shown in Table 2) as templates and then amplified by the established multiplex real-time PCR method compared with CSF (GB/T 27540-2011 2011),ASF (GB/T 18648-2020 2020),and APPV (CN108611442A 2018).
2.11.Detection of clinical samples by the multiplex real-time PCR
The established multiplex real-time PCR was used to detect 451 samples,including 246 samples from Xinjiang,198 clinical samples from Tianjin,six clinical samples from Hebei,and one clinical sample from Hunan,China.DNA or cDNAs of 451 clinical samples were extracted acrooding to a method for extracting nucleic acid bead method and then used as templates in the established multiplex real-time PCR to detect CSFV,ASFV,and APPV.
3.Results
3.1.Optimization of the singleplex real-time PCR reaction conditions
Optimizing by the matrix method and comparing the fluorescence intensity and Ctvalue between possible combinations,we found that the reaction system of 25 mL presents the optimized singleplex real-time PCR reaction condition (Table 5).The Singleplex Real-Time PCR Amplification Program on the LightCycler®480 II Instrument (Roche Life Sciences) was set: 95°C for 30 s;95°C for 10 s,annealing temperature and extension temperature at 58°C for 20 s,number of cycles for 45 cycles,collecting fluorescence and 40°C for 30 s,then run the program.
3.2.Optimization of the multiple real-time PCR reaction conditions
We first completed the optimized singleplex real-time PCR,and then the optimized the triple multiplex.The singleplex and multiple real-time PCRs of reaction conditions and reaction system were the same,but the primer and probe concentrations were different,as shown in Table 6.
3.3.Establishment of standard curves
Based on the optimal multiplex reaction system,this study optimized reaction conditions for three positive plasmids with dilutions of 1×107-1×101copies mL-1(three replicates for each gradient) and performed multiplex realtime PCR amplification.After the reaction,we drew a standard curve with the Ctvalue as the abscissa and the logarithm of the initial copy number concentration of the positive template as the ordinate.The results showed that the correlation coefficients (R2) of the three standard curves were all above 0.995,indicating that the numberof starting templates and Ctvalues showed a good linear relationship (Fig.1).
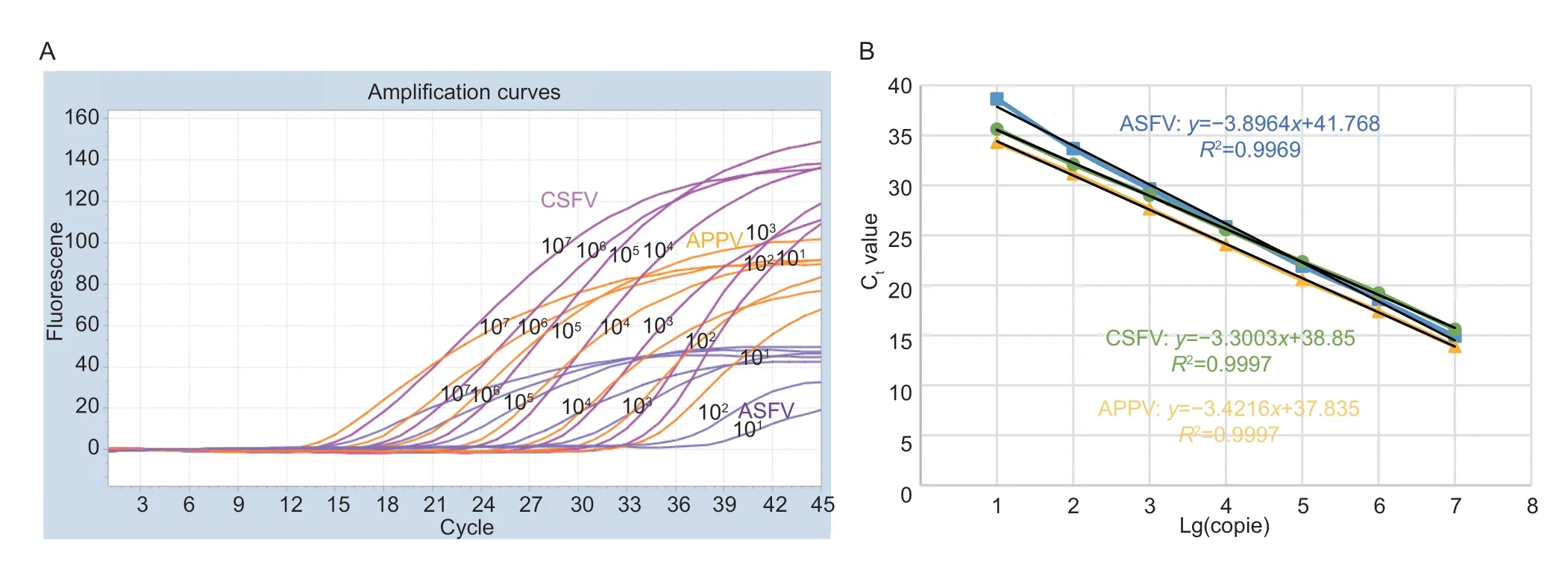
Fig.1 Amplification curves and standard curves were drawn by the multiplex real-time PCR.A,amplification curves of classical swine fever virus (CSFV),African swine fever virus (ASFV),and atypical porcine pestivirus (APPV) were drawn using each positive plasmid of concentrations from 1×107 to 1×101 copies mL-1.B,standard curves of CSFV,ASFV,and APPV were constructed using the mean of three replicate Ct values of each concentration.All amplification curves were conducted with Launch Software add-on for the Light Cycler 480 instrument.
3.4.Specificity of the multiplex real-time PCR assay
DNA and cDNA of CSFV,ASFV,and APPV were used as templates,and sterilized double distilled water was used as a negative control.The multiplex real-time PCR was used for specificity detection.The results indicated that the DNA or cDNA of CSFV,ASFV,and APPV showed specific amplification curves,the Ctvalue was less than 40 cycles,and the DNA or cDNA of other viruses and the negative control had no amplification curve.The established multiplex real-time PCR method demonstrated reliable specificity (Fig.2).Moreover,plasmids containing seven different genotypic ASFV p72 synthetic DNA were also amplified by the multiplex real-time PCR method.
3.5.Sensitivity of the multiplex real-time PCR assay
A total of 1 mL of each gradient of three positive plasmids 1×105-1×10-1copies mL-1was used as the template for amplification to obtain the multiplex real-time PCR-sensitive test result.The results showed that the detection limit of this method for CSFV,ASFV,and APPV was 1×100copy mL-1(Fig.2),and the crucial positive Ctvalues of CSFV,ASFV,and APPV were 38,39,and 38,respectively.
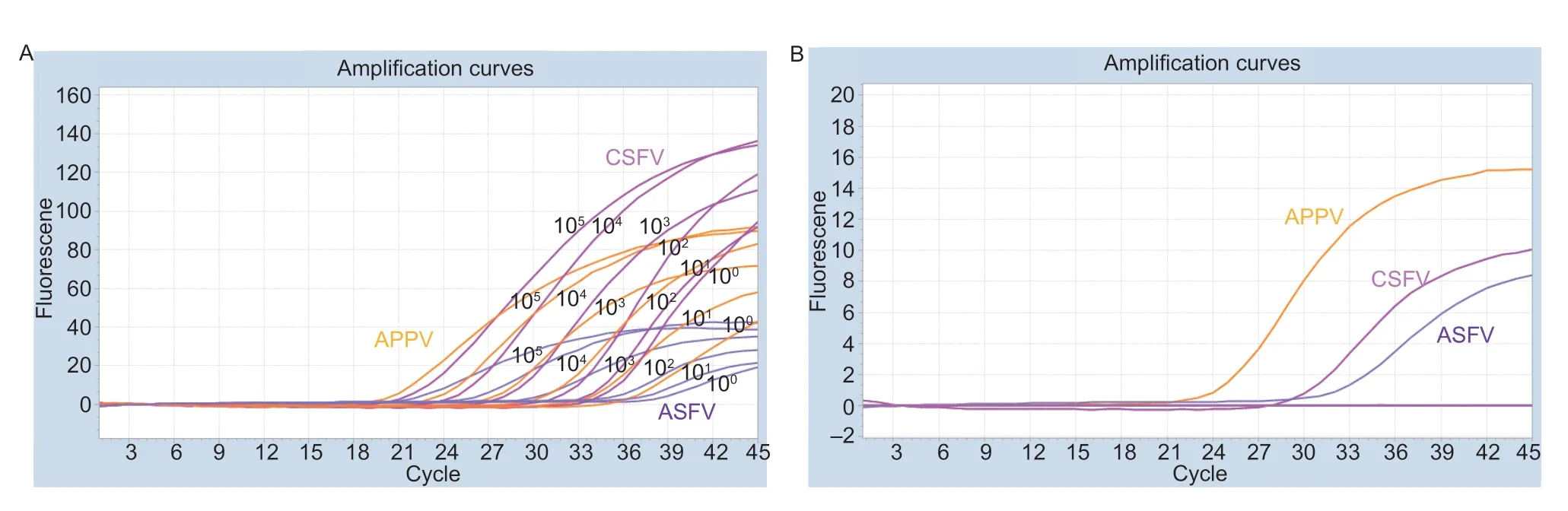
Fig.2 Specificity and sensitivity results of the multiplex real-time PCR.A,amplification curves of classical swine fever virus (CSFV),African swine fever virus (ASFV),and atypical porcine pestivirus (APPV) for each DNA/cDNA were detected by the multiplex realtime PCR,and negative control.B,amplification curves of each positive plasmid of concentrations from 1×105 to 1×10-1 copies mL-1 were detected by the multiplex real-time PCR.
3.6.Repeatability of the multiplex real-time PCR assay
We carried out reproducibility tests on the multiplex real-time PCR.The results showed that the variation coefficient of Ctvalues of the repeatability test within and between groups was less than 1% (Table 7),indicating that the method has good repeatability.
3.7.Comparison of the multiplex real-time PCR and other detection methods
In order to verify the reliability of the detection method,we used different methods to detect 22 samples (Table 2)by the multiplex real-time PCR,compared with national standard diagnostic and patented method assay for CSF (GB/T 27540-2011 2011),ASF (GB/T 18648-2020 2020),and APPV (CN108611442A 2018).The result of compliance was the same (100%) (Table 8).
3.8.Detection of clinical samples by the multiplex real-time PCR
This study collected 451 clinical samples from Xinjiang,Tianjin,Hebei,and Hunan.The results showed that the positive rates were CSFV (0.22%,1/451) and ASFV(1.3%,6/451).No APPV was detected positive in the 451 clinical samples.No coinfection was discovered in the positive samples (Table 9).
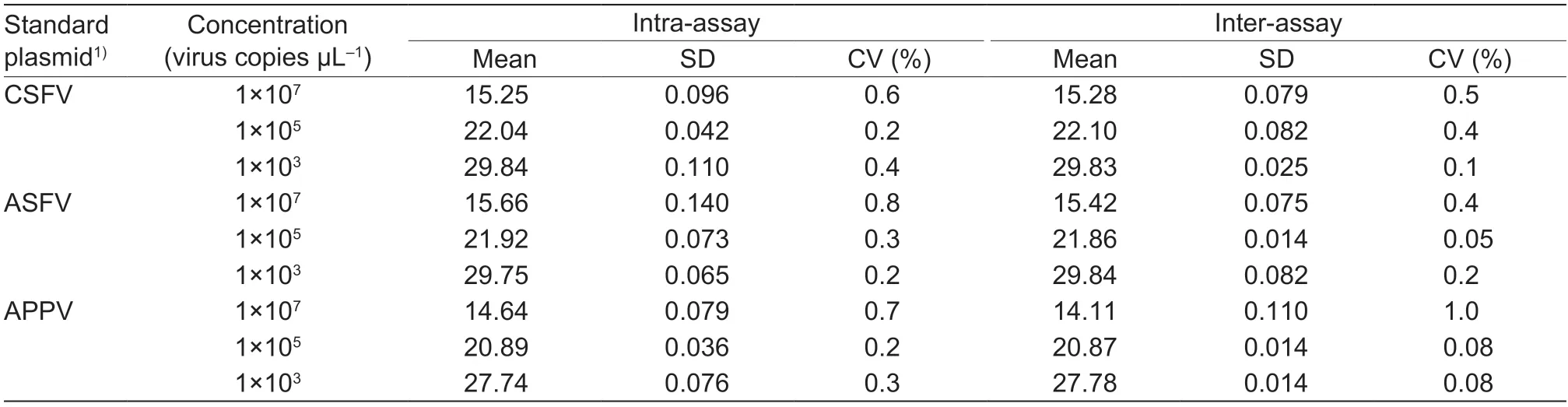
Table 7 Reproducibility assay of the multiplex real-time PCR
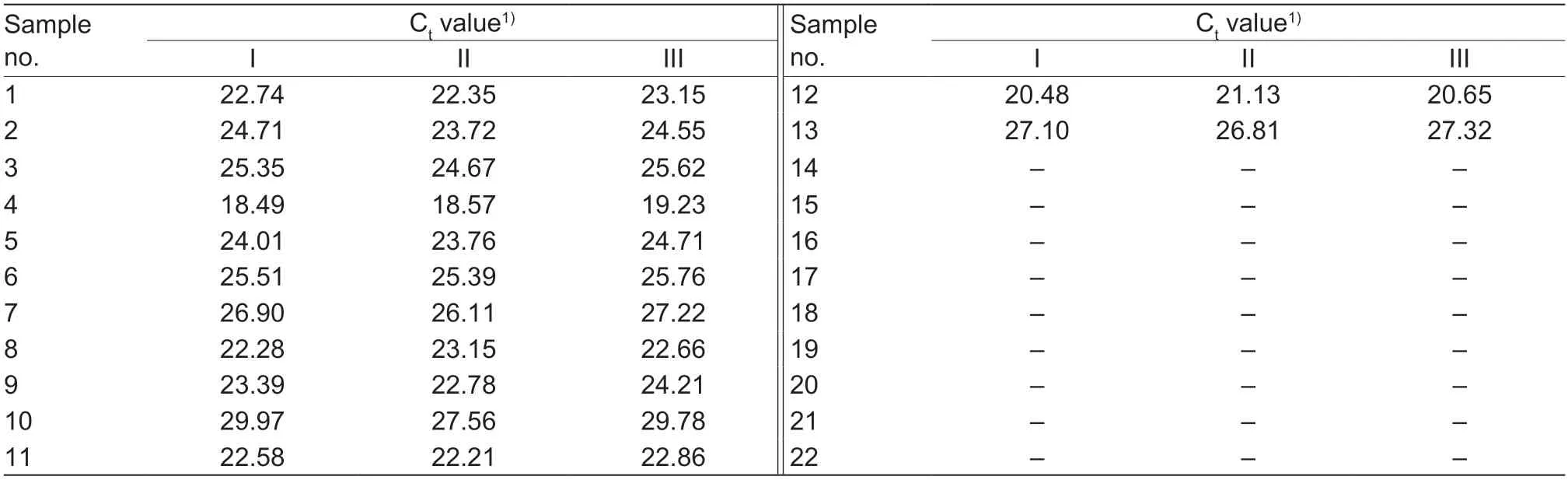
Table 8 Comparison of the multiplex real-time PCR from 22 samples
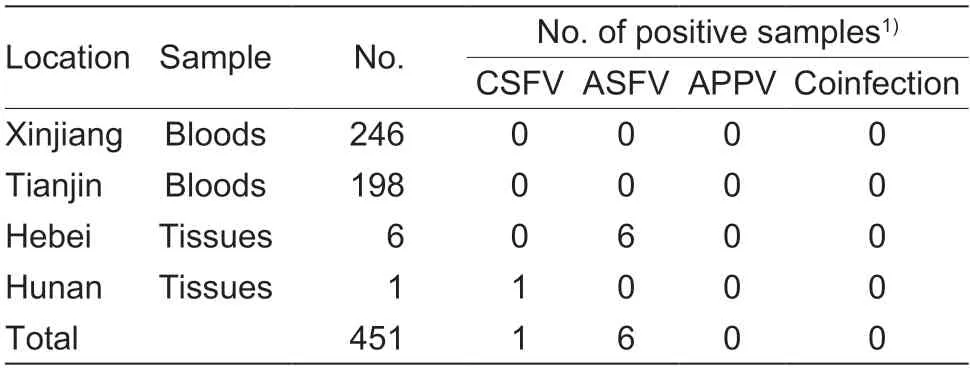
Table 9 Detection of clinical samples
4.Discussion
The C-strain vaccine is widely used in China to control the disease,and CSF occurs at least sporadically at present (Wanget al.2021;Weiet al.2021).ASF is a highly contagious global disease,and APPV is the emerging porcine pestivirus disease related to congenital tremor (CT) type A-II in piglets (Zhanget al.2018;Qiuet al.2021).CSFV,ASFV,and APPV seriously endanger the pig industry in China,and the clinical symptoms and pathological changes of these viruses are hard to distinguish.Therefore,a rapid,highly sensitive,specific,and accurate diagnosis of detection method is critical for the prevention and control of the diseases (Postelet al.2019;Zhou 2019;Liet al.2021).Virus isolation andserological testing are complicated and time-consuming,which require high laboratory conditions.However,real-time PCR is widely used in clinical detection,which has the advantages of sensitivity,high efficiency,quantification,and high throughput.
The multiplex PCR detection methods are difficult to establish because of the high requirements for the design of primers and probes.Moreover,when the probes are labeled with different fluorophores,the possibility of spectral overlap between probes may occur in multicolor fluorescent.When the fluorescence wavelength range crosses,a color compensation experiment is required.This study found that the sensitivity of the triple fluorescence PCR is lower than that of the singlplex PCR.On the one hand,this may be due to the interference of primers and probes during amplification;on the other hand,it may be the result of compensation between each fluorophore.In recent years,some researchers have developed and designed multiplex PCR or multiplex real-time PCR assays to simultaneously detect different pathogens.A panel of multiplex real-time PCR/RT-PCR assays composed of four sub-panels,with each detecting three common swine pathogens.In total,this assay can simultaneously detect and differentiate 12 common swine viruses with high specificity and sensitivity (Shiet al.2016).A multiplex reverse transcription PCR (mRT-PCR) was developed to simultaneously detect four RNA viruses in swine (Liuet al.2011).
It has been reported that mutant strains of ASFV have emerged in the field (Sunet al.2021),but theB646Lgene encoding p72 protein remains relatively conservative,with no changes in the primers and probe used in this study.Therefore,our assay is effective for detecting these ASFV mutant strains and other different genotypes.In this research,we have established the simultaneous detection of CSFV,ASFV,and APPV by a multiplex real-time PCR.After optimizing various reaction conditions (such as annealing temperature,primer and probe concentration,and the number of amplification cycles),a multiplex realtime PCR detection method with high sensitivity and good specificity was developed,which can simultaneously detect CSFV,ASFV,and APPV in one reaction system.The sensitivity of the assay is 1 copy mL-1,and the specificity is high.Nowadays,high specificity,sensitivity,time-saving,and cost-saving fluorescent PCR with the capability of detecting multiple pathogens in one reaction is in urgent need in laboratories for early diagnosis.This method not only enhances the detection ability but also reduces the workload and cost,which may be conducive to early clinical diagnosis and epidemiological research.
5.Conclusion
This study developed a multiplex real-time PCR detection method based on TaqMan probes to simultaneously detect CSFV,ASFV,and APPV in pigs.The detection limit of each pathogen can be as low as 1 copy mL-1,with high sensitivity,reasonable specificity,and reproducibility.The application of this method in clinical detection enhances detection ability and reduces the workload and cost,contributing to early clinical diagnosis and epidemiological research.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
The study has no ethical implications.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31872484) to Zhang Qianyi and the Non-profit Key Program of Veterinary Drug Industry from China Institute of Veterinary Drug Control (GY202011) to Xia Yingju.
杂志排行
Journal of Integrative Agriculture的其它文章
- A 314-bp SlNE insertion in the ZNF2 promoter region may act as a repressor related to regulation of fat deposition in pigs
- An optimized protocol using Steedman’s wax for high-sensitivity RNA in situ hybridization in shoot apical meristems and flower buds of cucumber
- Characterization of subunits encoded by SnRK1 and dissection of combinations among these subunits in sorghum (Sorghum bicolor L.)
- The role of time preferences in contract breach: Evidence from Chinese poultry farmers participating in contract farming
- Optimal design of culling compensation policy under the African swine fever — Based on simulations of typical pig farms in China
- Drip fertigation and plant hedgerows significantly reduce nitrogen and phosphorus losses and maintain high fruit yields in intensive orchards
