Characterization of the chemosensory protein EforCSP3 and its potential involvement in host location by Encarsia formosa
2023-02-03WANGKeHEYanyanZHANGYoujunGUOZhaojiangXIEWenWUQingjunWANGShaoli
WANG Ke,HE Yan-yan,ZHANG You-jun,GUO Zhao-jiang,XIE Wen,WU Qing-jun,WANG Shao-li
Institute of Vegetables and Flowers,Chinese Academy of Agricultural Sciences,Beijing 100081,P.R.China
Abstract Chemosensory proteins (CSPs) perform several functions in insects.This study performed the gene expression,ligand-binding,and molecular docking assays on the EforCSP3 identified in the parasitoid wasp Encarsia formosa,to determine whether EforCSP3 functions in olfaction,especially in host location and host preference.The results showed that EforCSP3 was highly expressed in the female head,and its relative expression was much higher in adults than in other developmental stages.The fluorescence binding assays suggested that the EforCSP3 exhibited high binding affinities to a wide range of host-related volatiles,among which dibutyl phthalate,1-octene,β-elemene,and tridecane had the strongest binding affinity with EforCSP3,besides α-humulene and β-myrcene,and should be assessed as potential attractants.Protein structure modeling and molecular docking predicted the amino acid residues of EforCSP3 possibly involved in volatile binding.α-Humulene and β-myrcene attracted E.formosa in a previous study and exhibited strong binding affinities with EforCSP3 in the current study.In conclusion,EforCSP3 may be involved in semiochemical reception by E.formosa.
Keywords: Encarsia formosa,chemosensory protein,expression profiles,fluorescence binding assay,molecular docking
1.Introduction
When attacked by herbivorous insects,plants may release volatile compounds to attract natural enemies of the herbivores and thereby protect themselves(Kessler and Baldwin 2001;Schumanet al.2012).Such natural enemies are usually insects and often perform physiological activities,such as foraging,host searching,and oviposition,based on the chemical odors through their olfactory system (Kaissling 2001;Swansonet al.2009).The sensation and transmission of insect olfaction are mediated by olfactory proteins,causing insect physiological and behavioral responses (Leal 2013).Olfactory proteins include chemosensory proteins (CSPs),odorant-binding proteins (OBPs),odorant receptors(OR),sensory neuron-membrane proteins,and odorantdegrading enzymes (Younaset al.2018).Among these proteins,CSPs are also believed to be critical for the primary stage of chemoreception and signal transmission in insects except for OBPs (Pelosiet al.2018;Zhanget al.2020;Zhouet al.2020).
CSPs are small soluble proteins that occur in the insect lymph fluid.They are highly conserved and always possess hydrophobic binding pockets formed by α-helical domains (Tegoniet al.2004).Their main function is to combine with odorant molecules and then transport those molecules to receptors (Campanacciet al.2003;Pelosiet al.2006;Leal 2013).In insects,CSPs are reported to be involved in different functions,including the pheromone recognition ofSchistocerca gregaria(Angeliet al.1999),embryonic development ofApis mellifera(Ozakiet al.2008),insecticide resistance inBemisia tabaciandAnopheles gambiae(Liuet al.2014,2016),reproduction ofB.tabaci(Zenget al.2020),and host searching ofRhopalosiphum padi(Penget al.2021).Although CSPs may have many functions,their main function appears to involve chemoreception,given that most CSPs are highly expressed in antennae of pests and parasitoids (Liuet al.2012).Evidence that CSPs function in host location is mainly based on the analyses of gene expression and gene silencing,and on fluorescence binding assays (Guet al.2012;Zhanget al.2014,2020).In the herbivorous mothCnaphalocrocis medinalis,CmedCSP33 forms a stable complex with the host plant volatiles nerolidol and β-ionone (Duanet al.2019).In another example,CSP2 mediates the response ofB.tabacito plant volatiles and is involved in the whitefly’s feeding and oviposition preferences;the CSP2 protein inB.tabacican bind to the plant volatiles,including homoterpene (E)-3,8-dimethyl-1,4,7-nonatriene (DMNT),which can influence host selection and oviposition ofB.tabaci(Liet al.2021).
The parasitoidEncarsia formosaGahan has been widely applied to reduce damage caused byB.tabaciand other whiteflies on vegetables and ornamental plants grown in greenhouses.Its chemosensory behaviour in response toB.tabacihas been described (Liuet al.2014),and its selection between two biotypes ofB.tabacidiffered depending on whether the whitefly-infested plants were infected with a begomovirus;the latter effect was closely related to elevated levels of some host plant volatiles (Liuet al.2017).Although these results indicate that olfactory recognition is involved in host location and preference byE.formosa,the mechanism by whichE.formosarecognizes chemical volatiles remains unknown.In previous studies,10 CSPs were identified based on the antennal transcriptome ofE.formosa(Heet al.2020),among which the relative expression ofEforCSP3was found to be much higher than otherCSPgenes.It is predicted that thisEforCSP3gene might be involved in host location through the perception of the infested plant volatiles.
This study identified and assessed the relative expression of theEforCSP3gene inE.formosabased on its antennal transcriptomic data.After determining thatEforCSP3expression is much higher in the head than in other body tissues and is higher in adults than in other stages,we analyzed the ligand-binding affinity of EforCSP3 and characterized its structural properties by molecular docking.These results increase the understanding of the olfactory mechanisms underlying host selection byE.formosaand could contribute to the use of the wasp for biological control in the future.
2.Materials and methods
2.1.Insect rearing and sampling
A culture ofE.formosawas provided by the Research Center for Beneficial Insects,Institute of Plant Protection,Shandong Academy of Agricultural Sciences in China.EmergedE.formosawere reared onB.tabacinymphs that were feeding on tomato plants,Solanum lycopersicum,in a growth chamber ((26±2)°C,(70±5)%relative humidity,and 14 h L:10 h D).TheE.formosadevelopmental stages included pupae and adult females at 6,12,and 24 h after emergence,with approximately 100 individuals as a sample (one biological replicate) for each developmental stage.For assessment of different tissues,1 000 female wasps emerged for 6 h were cut into two parts: the antennae (head) and a mixture of other body parts (thorax,wings,abdomen,and legs),with each part as one replicate.Each treatment was represented by three biological replicates.The samples were immediately frozen in liquid nitrogen and stored at -80°C for subsequent analysis.
2.2.RNA isolation and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen,Carlsbad,CA,USA) according to the manufacturer’s instructions.The integrity and purity of extracted RNA were assessed by electrophoresis,and RNA quantity was determined using a Nanodrop 2000 (Thermo Scientific,Wilmington,DE,USA).The first-strand cDNA was synthesized using 1 μg of total RNA with the Prime Script®RT Reagent Kit (TaKaRa Biotech,Kyoto,Japan) following the manufacturer’s recommendations;the first-strand cDNA was stored at -20°C.
2.3.Gene identification and phylogenetic tree construction
Based on the 10 CSPs genes fromE.formosaantennal transcriptomic data (Heet al.2020),theEforCSP3gene ofE.formosawas annotated and submitted to GenBank Database in National Center for Biotechnology Information(NCBI) under the accession no.MN830262.SignalPV5.0(http://www.cbs.dtu.dk/services/SignalP/) was used to predict the putative N-terminal signal peptides and cleavage sites ofEforCSP3.The occurrence of α-helices and molecular weight were predicted using ExPASy(https://www.expasy.org/).The homologous genes ofEforCSP3from Hymenoptera insect orders were identified using the Basic Local Alignment Search Tool (BLAST)in NCBI.MEGA X was used for the construction of a phylogenetic tree by the maximum likelihood method based on the bootstrap procedure with 1 000 replicates(Caspermeyer 2018).Circular phylogenetic trees were then prepared and colored according to taxonomy with iTOL (https://itol.embl.de).
2.4.Gene expression by real-time quantitative PCR(qRT-PCR)
qRT-PCR with an ABI Prism 7500 real-time PCR System(Applied Biosystems,Foster City,CA,USA) was used to compare the relative expression ofEforCSP3in the antenna (head)vs.the body (including the thorax,wings,abdomen,and legs),and in pupaevs.adults(at 6,12,and 24 h after eclosion).The forward primer(5´-AGGTCGTAGTAACTTCATGCAA-3´) and reverse primer (5´-TGGTCTTGACAACGAGCTCT-3´) were used for the qRT-PCR procedure.Each 20 μL reaction contained 10 μL of 2× FastFire qPCR PreMix,0.4 μL of 50× ROX reference dye,0.6 μL of forward primer (10 μmol L-1),0.6 μL of reverse primer (10 μmol L-1),1.0 μL of cDNA (300 ng μL-1),and 7.4 μL of RNase-free ddH2O according to the instructions provided by the FastFire qPCR PreMix (SYBR Green) Kit (Tiangen,Beijing,China).The qRT-PCR program was as follows: polymerase activation at 95°C for 1 min,followed by 40 cycles of denaturation at 95°C for 5 s,annealing at 60°C for 10 s,and elongation at 72°C for 15 s.For melting curve analysis,a dissociation step cycle was automatically added.Relative quantification was performed according to the 2-ΔΔCTmethod,and the data were normalized to the reference genes,β-actinandGADPH,according to Heet al.(2020).Each reaction was performed three times with different biological samples.
2.5.Construction and purification of recombinant protein
The EforCSP3 was amplified by PCR using a forward primer (5´-CTAAGCTTATAATCAAGCAGTCCTCT-3´)containing aHindIII-restriction site and a reverse primer (5´-CTGAATTCGACGAAGAGGCCAGACGA-3´)containing anEcoRI-restriction site.The ligation of the PCR product was performed using a vector pTOPO-T(Aidlab),and the ligation product was subsequently transformed into DH5α cells ofEscherichia coli.The positive clones were picked,cultured in Luria-Bertani(LB) medium (50 μg mL-1kanamycin),and sequenced.Restriction enzymes (EcoRI andHindIII) were used to digest the target fragment,which was then ligated into the pET-30a (+) expression vector.The recombinant plasmid was subsequently transformed intoE.coliDH5α cells.After sequencing,the confirmed recombinant plasmid was transformed intoE.coliBL21 (DE3) cells.A single clone was cultured in LB medium,and the positive colonies were confirmed by sequencing.
Expression and purification were performed using a positive clone cultured in 10 mL of LB medium (50 μg mL-1kanamycin).The culture was diluted to 1 L with LB medium and incubated until an optical density (OD600)of 0.4-0.6 was obtained.For the induction of protein expression,isopropyl-beta D-thiogalactopyranoside(IPTG;0.1 mmol L-1) was added,and the mixture was cultured on a shaking table at 16°C for 10 h.After that,purification was conducted in a nickle (Ni) affinity chromatography column (His-Tagged Protein Purification Kit,CWBIO,China).The His-tag was removed from the eluted protein by adding recombinant bovine enterokinase at 26°C for 15 h.To obtain tag-free protein,the digested protein was passed through the Ni affinity chromatography column again.The purified protein was desaltedviaextensive dialysis,and the size and purity of the recombinant protein were confirmed by 4-20% SDSPAGE (SurePAGE,GenSript,China) analysis.
2.6.Fluorescence binding assay
Tomato volatiles were previously reported to have different levels in healthyvs.TYLCV-infected tomato plants,and some were proven to be critical cues in host selection byE.formosa(Liuet al.2017).Therefore,a total of 20 candidate ligands including monoterpenes,sesquiterpenes,aromatic compounds,alkanes,aldehydes,esters,and alcohols (Table 1),were used to determine the binding ability of EforCSP3 using N-phenyl-1-naphthylamine (1-NPN),a fluorescent probe.The 1-NPN was diluted with methanol to obtain a 1 mmol L-1stock solution.The aliquots of 1-NPN were added into the 2 μmol L-1stock solution EforCSP3 protein that was diluted with PBS buffer (50 mmol L-1,pH 7.4).The potential ligands were prepared in a similar manner as 1-NPN.To measure the affinity of 1-NPN and EforCSP3,the 2 μmol L-1EforCSP3 protein solution was titrated with 1 mmol L-11-NPN to a final concentration(2-20 μmol L-1).The mixture of 1-NPN/EforCSP3was excited at 285 nm wavelength,and the emission spectra ranging from 290 to 530 nm were recorded using a spectrofluorophotometer (970-CRT,Lengguang,Shanghai,China).Three independent replicates were performed in this experiment.
2.7.3D modeling and molecular docking
The 3D structure of the EforCSP3 protein was modeled with the online I-TASSER Program (Royet al.2010;Yanget al.2015).The final 3D model was assessed with online Structure Analysis and Verification Servers(http://services.mbi.ucla.edu/SAVES/),including Procheck,Verify_3D,and ERRAT.Molecular docking was conducted to determine the binding of ligands using Autodock (version 1.5.6),which was used to generate the docking input files (Morriset al.2009;Forliet al.2016).The search grids (binding pockets) were identified by the EforCSP3 model protein.To increase the docking accuracy,the value of exhaustiveness was set at 20.For Autodock,the default parameters were used as described in the Autodock manual unless specified otherwise.The top-ranked conformation,in combination with the docking score,was subjected to visual analysis using PyMOL(Open-source version 2.4.0) (https://pymol.org/).
2.8.Data analysis
The EforCSP3 binding affinities (Ki) for each compound were calculated using the corresponding IC50values(concentration of a competitor that reduced the initial fluorescence intensity by half) based on the following equation:

where [1-NPN] is the free concentration of 1-NPN,andK1-NPNis the dissociation constant of the EforCSP3/1-NPN complex.The comparative analysis ofEforCSP3expression among different developmental stages was conducted using one-way ANOVA followed by Tukey’s HSD test and an independentt-test that analyzed headvs.body tissue.All data are presented as the mean±SE,and the SPSS (v.25.0) was used for data analysis.
3.Results
3.1.Characterization,sequence analysis,and phylogenetic analysis of EforCSP3
After theEforCSP3gene was obtained from an antenna transcriptome ofE.formosa,a full-length cDNA encoding was cloned and verified by sequence analysis.The open reading frame (GenBank no.MN830262.1) consisted of 123 amino acid residues encoded by 372 nucleotides,with a molecular weight of 14.11 kDa and an isoelectric point of 4.80.The EforCSP3 protein had signal peptide of 17 amino acid residues at the N-terminus (Fig.1).An alignment of amino acid sequences indicated that the CSPs of other hymenopteran species shared 36-61%sequence identity withEforCSP3(Appendix A).TheEforCSP3gene has four conserved cysteines and a sequence motif of C-X6-C-X19-C-X2-C.
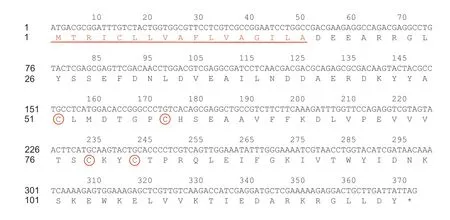
Fig.1 Nucleotide and deduced amino acid sequence of EforCSP3.Four conserved cysteines of EforCSP3 are shown in red circles.The underlined red sequence indicates 17 predicted signal peptides.
To determine the possible functions of EforCSP3,a phylogenetic tree was constructed based on the amino acid sequences of EforCSP3 and 117 CSP genes of Hymenoptera insects from the NCBI Database.The alignment analysis showed that the percentage of identical amino acids was approximately 11.57-57.98%.According to the morphological characteristics of the phylogeny,EforCSP3 clustered with CcunCSP1 with 57.98% amino acid identity (Fig.2).
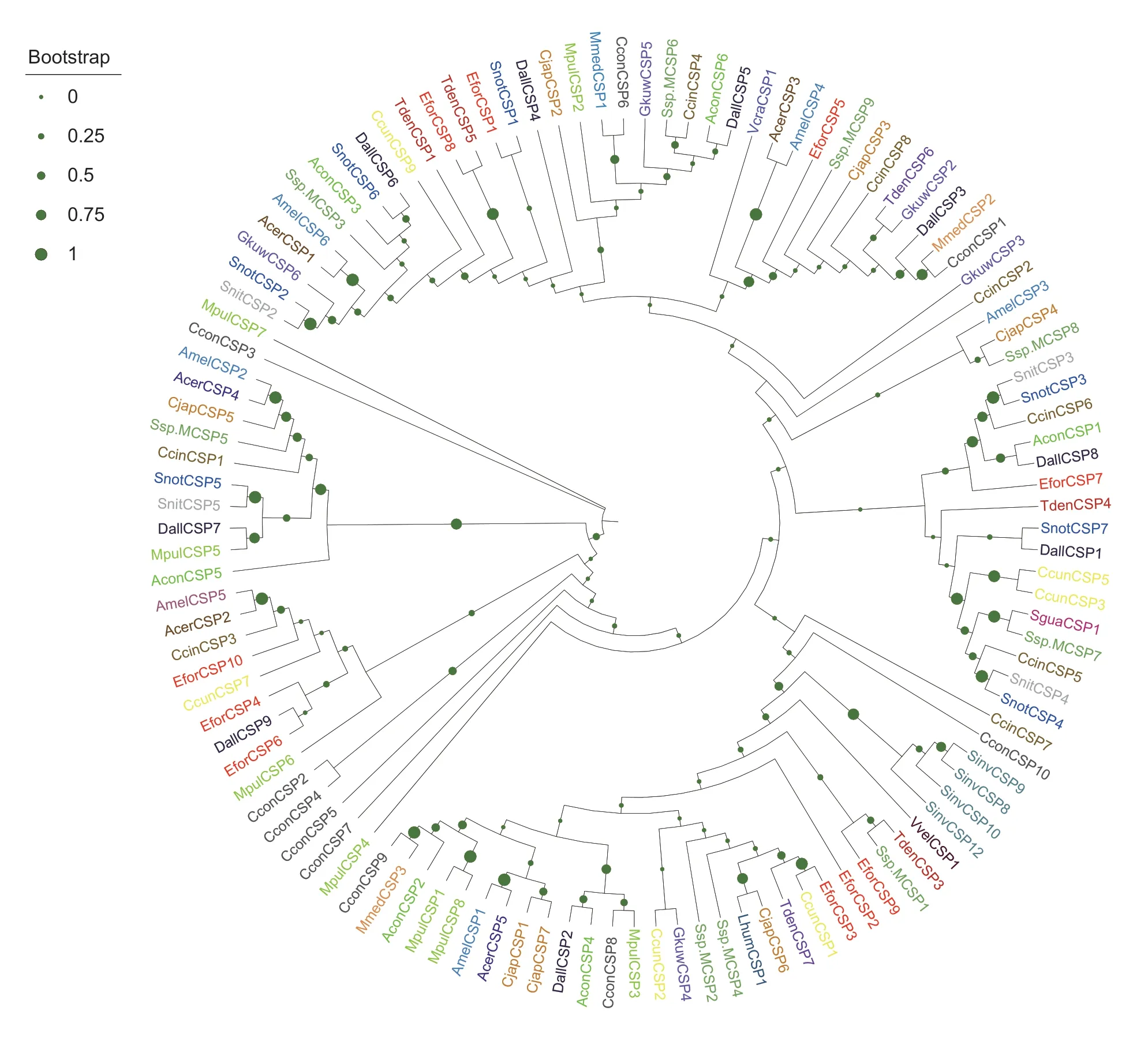
Fig.2 Phylogenetic tree of EforCSP3 with 117 chemosensory proteins from Hymenoptera insect species.EforCSP3 is marked with a red pentagram.The chemosensory proteins originated from Vespa crabro (Vcra),Apis cerana cerana (Acer),Camponotus japonicus (Cjap),Apis mellifera (Amel),Encarsia formosa (Efor),Meteorus pulchricornis (Mpul),Microplitis mediator (Mmed),Linepithema humile (Lhum),Trichogramma dendrolimi (Tden),Sclerodermus guani (Sgua),Sclerodermus sp.MQW-2015 (Ssp.M),Solenopsis invicta (Sinv),Gregopimpla kuwanae (Gkuw),Cotesia congregate (Ccon),Sirex nitobei (Snit),Aulacocentrum confusum(Acon),Chouioia cunea (Ccun),Diachasma alloeum (Dall),Vespa velutina (Vvel) and Cephus cinctus (Ccin).
3.2.Expression profiles of EforCSP3 in different tissues and developmental stages
AmongE.formosatissues,EforCSP3expression was substantially higher in the head than in the abdomen(P<0.001) (Fig.3-A),indicatingEforCSP3may participate in chemoreception based on its expression characteristics.AmongE.formosadevelopmental stages,EforCSP3expression was much higher in adults than in pupae;expression was the highest in recently emerged adults (6 h) and then gradually decreased with time after emergence (Fig.3-B).

Fig.3 Expression profiles of EforCSP3 in Encarsia formosa.A,expression level in the head and abdomen.Values (mean±SE,n=3)with different letters are significant at P<0.001 according to a t-test.B,expression level in pupae and in adults at 6,12,and 24 h after adult emergence.Values (mean±SE,n=3) with different letters are significantly different at P<0.001 according to Tukey’s test.
3.3.Fluorescence binding assay
After the signal peptide and terminator were removed,the recombinant EforCSP3 was successfully induced and expressed inE.coli;it was mainly expressed in the supernatant,as shown by SDS-PAGE (4-20%),and the molecular weight was 14.10 kDa (Fig.4).The purified EforCSP3 was used in fluorescence binding experiments to assess the binding of EforCSP3 to various ligands.
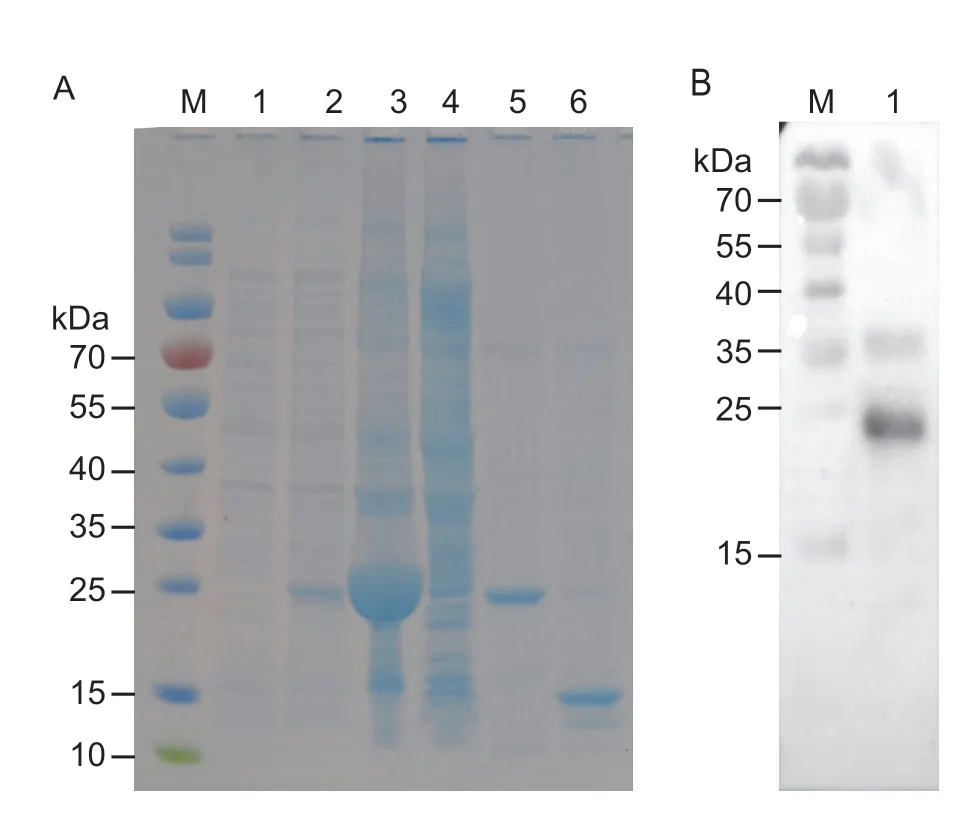
Fig.4 SDS-PAGE analysis for the expression and purification of EforCSP3.A,Western blot analysis of the expression of EforCSP3.M,protein marker;lane 1,non-induced PET30a/EforCSP3 before IPTG induction;lane 2,IPTG-induced PET30a/EforCSP3;lane 3,supernatant of induced PET30a/EforCSP3;lane 4,sediment of induced PET30a/EforCSP3;lane 5,purified EforCSP3 with His-tags;lane 6,purified protein of pET30a/EforCSP3 without His-tags.B,purification of EforCSP3.M,protein marker;lane 1,fusion protein.
To determine the relative affinities of EforCSP3 with the different quantities of 20 volatiles produced from both healthy and TYLCV-infected tomato plants,fluorescence binding assays were conducted.The results showed that EforCSP3 was strongly bound to fluorescent probe 1-NPN.According to the changes in maximal fluorescence intensity (Appendix B),the dissociation constant (Kd) of the EforCSP3/1-NPN complex calculated from Scatchard plots was 2.48 μmol L-1at pH 7.4.A saturation and linear Scatchard plot suggested the existence of a single binding site in the protein with no allosteric effects (Appendix B).
The values of IC50andKiof ligands were calculated and are shown in Table 1.The fluorescence binding assay showed that dibutyl phthalate had the strongest binding affinity with aKivalue of 4.14 μmol L-1,while 1-octene,β-elemene,tridecane,α-humulene,tetradecane,and β-myrcene also had very strong binding affinities withKi<6 μmol L-1to EforCSP3.Moreover,all of the alkanes,esters,and sesquiterpenes could bind well with EforCSP3 (Table 1),indicating that EforCSP3 may function as a relatively broad binding protein in volatile recognition.
3.4.Homology modeling and molecular docking
To identify the key binding sites and crucial amino acid residues involved in binding,we used computational procedures to model the 3D structure of EforCSP3.According to the Protein Data Bank (PDB),EforCSP3 protein has <30% homology with known templates;the structure was therefore predicted using the online I-TASSER Program (https://zhanglab.ccmb.med.umich.edu/I-TASSER/).The 3D structure of EforCSP3 was modeled with SgreCSP4 (PDB ID: 2GVS) as a template(Fig.5),and the model was selected based on the root mean square deviation (RMSD) value (0.386 Å).All of the model qualities had G-factor values>-0.5,indicating that the distribution of torsion angles and covalent geometries of the modeled EforCSP3 protein were reasonable,and the 3D modeling was reliable.The protein structure of EforCSP3 included six typical α helices with α-1 (Asp34-Leu39),α-2 (Asp42-Leu52),α-3 (Ser60-Val75),α-4 (Pro83-Asp98),α-5 (Ser101-Thr111),and α-6 (Lys117-Leu120),and an internal cavity,defined as a hydrophobic binding cavity,which are key features of insect CSPs.
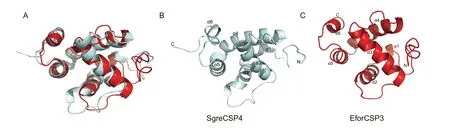
Fig.5 Structural modeling of EforCSP3.A,superimposed structure of EforCSP3 and the template.B,3D structure of SgreCSP4 used as a template.C,3D structure of EforCSP3.
Docking results showed strong binding sites of EforCSP3 with corresponding ligands,and the binding energy between EforCSP3 and each ligand was then calculated (Table 2).The results showed that all docking binding energies were negative,and that the distances of potential interaction residues were <4 Å (Fig.6).
EforCSP3 had a cavity wall composed of several residues such as Ala4,Asp37,Glu44,Ser60,Phe48,and Asp51 (Table 2 and Fig.6).Residues Ala4,Arg5,Leu8,Tyr9,Asp51,and Gln68 were critical sites in binding monoterpene and sesquiterpenes,and hydrogen bonds were formed between dibutyl phthalate and Arg5 residue.The interactions between 10 amino acid residues with alkane compounds,10 amino acid residues with monoterpene compounds,and 8-12 amino acid residues with sesquiterpene compounds were especially closely(Table 2 and Fig.6).The remaining ligands could also interact with amino acid residues of EforCSP3.
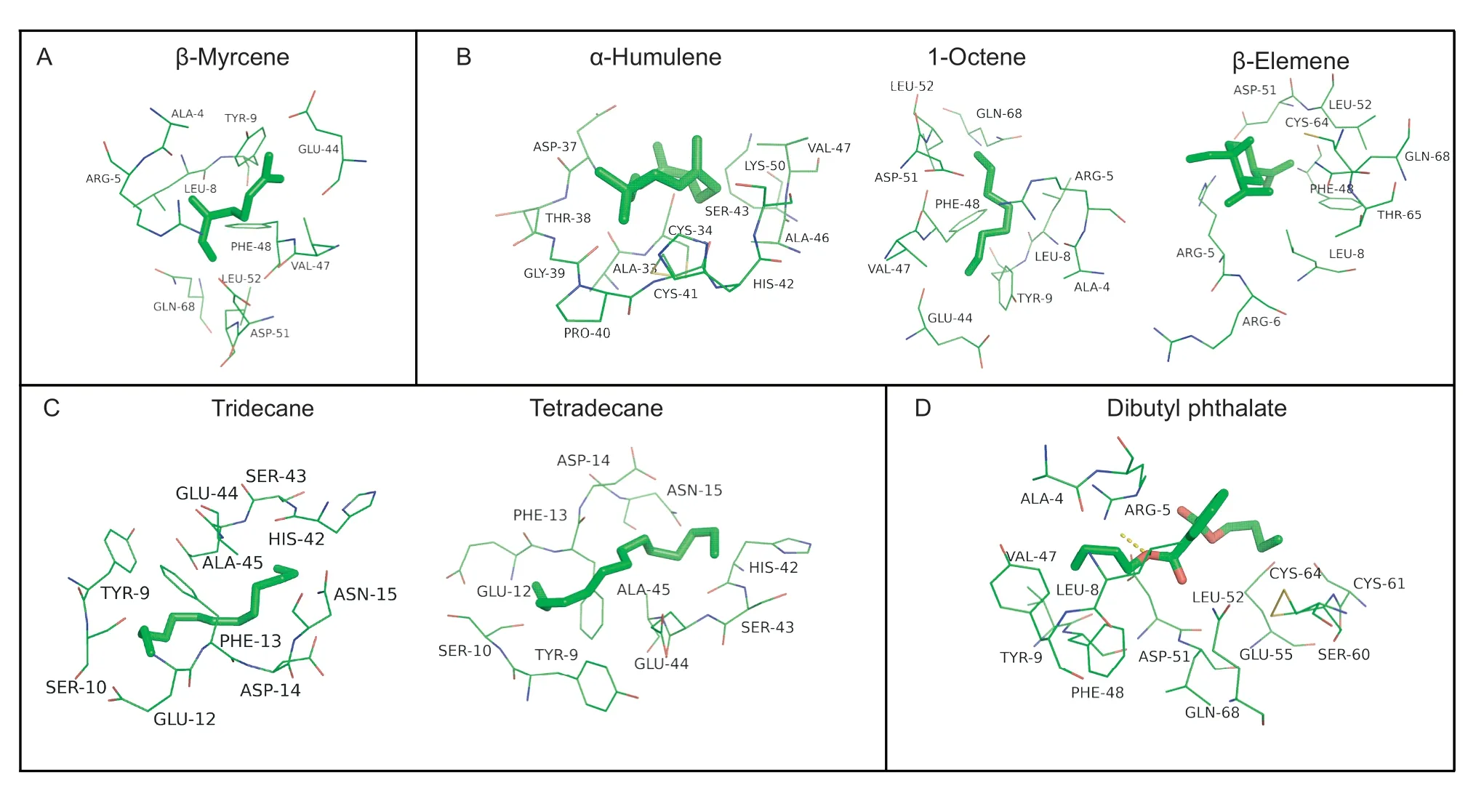
Fig.6 Docked conformation of ligands (A-D) into the putative binding pocket on EforCSP3.A,monoterpene.B,sesquiterpene.C,alkane.D,ester.Dashed lines in the D represent the hydrogen bonds.

Table 2 Molecular docking results of tested ligands
4.Discussion
Some CSP genes have been identified in parasitic wasps,includingMicroplitis mediator,Sclerodermussp.,Trichogramma dendrolimi,andChouioia cunea.Previous studies showed that CSPs might have multiple functions,including olfactory and non-olfactory perception (Zhanget al.2009,2015,2016;Zhaoet al.2016).In the present study,EforCSP3,which was recently identified in an antennal transcriptome analysis ofE.formosa(Heet al.2020),was found to encode a protein with the typical characteristics of CSPs (Pelosiet al.2006) and to be closely associated withCSPsin other hymenopterans.Analysis of the relative expression profiles of CSP genes could help understand their possible functions in insects.Many CSP genes expressed at high levels in antennae or antennae-specific were reported to perceive and recognize sex pheromones or general odorants (Calvelloet al.2005;Dippelet al.2014;Penget al.2017;Younaset al.2018;Wuet al.2019).Knockdown ofCSP4orCSP5,which are highly expressed in the antennae ofR.padi,significantly decreased the insect’s attraction to octanal,indicating that CSPs are involved in theR.padihost-location behavior (Penget al.2021).These studies showed that CSPs expressed in antennae possibly relate to the olfactory selection or host location.In the present study,the relative expression ofEforCSP3was significantly higher in adults than that in pupae,with the highest in adults 6 h after emergence,when theE.formosawasps conduct host searching,selection,and parasitism (Burger 2002),which is similar to a previous study (Heet al.2020).Furthermore,theEforCSP3was expressed significantly higher in the head than in the other body parts,also suggesting thatEforCSP3might be involved in the host location byE.formosa(Liet al.2019).Except for the perception for volatiles,other functions of CSP genes have also been identified,including the regeneration of the foot inPeriplaneta americana(Kitabayashiet al.1998),reproduction,feeding and oviposition preference inB.tabaci(Zenget al.2020;Liet al.2021).In addition,the SlituCSP18 protein inSpodoptera lituramight help mediate the interaction of the insect’s olfactory system and the pesticide chlorpyrifos (Linet al.2018).
Fluorescence-binding assays are commonly used to test the binding affinities of CSPs with small molecules.Generally,theKivalue is negatively correlated with the binding ability;the target protein binding glands havingKiless than 10 μmol L-1could be regarded to have strong binding affinities in most literatures (Younaset al.2018;Liet al.2019;Wang Z Het al.2021).From previous studies,theKiis not a fixed standard for binding affinity,and some reports adoptedKi<20 μmol L-1(Zhouet al.2020),Ki<5 μmol L-1(Wariset al.2020;Wang Qet al.2021) as strong binding,or others regardless of theKivalues.In this present study,Kivalues less than 10 μmol L-1were found in most tested compounds with EforCSP3,showing that EforCSP3 has wide binding properties.Especially,1-octene,β-elemene,tridecane,α-humulene,tetradecane,β-myrcene,and dibutyl phthalate had much stronger binding affinities withKi<6 μmol L-1to EforCSP3,compared to the rest volatiles (6 μmol L-1<Ki<12 μmol L-1).Our results were consistent with the previous finding that α-humulene and β-myrcene could attractE.formosain an olfactometer assay (Liuet al.2017).Therefore,we could infer that the EforCSP3 inE.formosacontributes to the location of hostsviaolfaction,even though it is not sure whether the other compounds having high binding affinities to EforCSP3,except α-humulene and β-myrcene,would attract theE.formosawasp currently.Meanwhile,the EforCSP3 ofE.formosacould bind wide volatiles in this study,which is in accord with other CSPs in Liet al.(2021) and Penget al.(2021);the CmedCSP1 and CmedCSP2 inCnaphalocrocis medinaliswere shown to have high binding affinities to a wide range of hostrelated semiochemicals (Zenget al.2018).Based on the data obtained from the ligand-binding experiment in this study,it is believed that the volatiles showing high binding affinity with EforCSP3 could possibly serve as a valuable resource for classifying attractants forE.formosaand pave the way for improving the biological control of whiteflies.
Results of ligand binding were further validated by 3D modeling and molecular ligand docking.A 3D model indicated that EforCSP3 has a structure characteristic of CSPs,i.e.,EforCSP3 consists of six α-helices and four conserved cysteines that form a binding pocket(Zhanget al.2012).The docking results in this study demonstrated that several amino acid residues of EforCSP3 could result in strong hydrophobic interactions with ligands,most likely because the binding pocket of EforCSP3 contains hydrophobic residues,which is consistent with Tianet al.(2016).Similar related reports,including Tyr26 for the binding of 12-bromo-dodecanol with MbraCSPA6 (Campanacciet al.2001),Y11 for the binding of BtCSP2 to DMNT (Liet al.2021),and Glu122 and Lys121 for the binding of MsepCSP8 with volatile compounds (Younaset al.2018),have been documented.In the present study,some amino acid residues (Ala4,Asp37,Glu44,Ser60,Phe48,and Asp51) could bind to ligands in the binding pocket of EforCSP3.More specifically,β-myrcene could bind to the sites of Glu-44,Val-47,Phe-48,Asp-51,and Leu-52 in the binding pocket of EforCSP3,which indicated thatE.formosapossibly recognize β-myrceneviathese amino acid residues of EforCSP3.In addition,hydrogen bonding between residue (Arg5) and ligands also supported some important amino acid residues for the interaction between EforCSP3 and ligands.Overall,the molecular docking showed that several amino acid residues with specific recognition site are involved in ligand binding by EforCSP3 ofE.formosa.Further studies are needed to determine whether potential attractants,such as 1-octene,β-elemene,tridecane,α-humulene,tetradecane,β-myrcene,and dibutyl phthalate,can attractE.formosain the field and confirm which amino acid residue is essential for the binding to a particular attractant through functional mutation experiments.The RNAi and behavioral assays are also required to clarify the physiological functions of the binding ligands from plants and the amino acid residues of EforCSP3.
5.Conclusion
TheEforCSP3was highly expressed in the head ofE.formosaadult females,and its expression was significantly higher in adults than that in pupae.Binding assays indicated that EforCSP3 binds to several whiteflyinfested plant volatiles,with a high affinity to seven volatiles.Furthermore,3D modeling and molecular docking predicted that some key amino acid residues of EforCSP3 could help bind the volatiles.In addition to the previous finding that α-humulene and β-myrcene could attractE.formosa,the current study found that EforCSP3 might contribute toE.formosalocation of hosts.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31772172),the earmarked fund for China Agriculture Research System (CARS-25) and the Beijing Key Laboratory for Pest Control and Sustainable Cultivation of Vegetables.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- A 314-bp SlNE insertion in the ZNF2 promoter region may act as a repressor related to regulation of fat deposition in pigs
- An optimized protocol using Steedman’s wax for high-sensitivity RNA in situ hybridization in shoot apical meristems and flower buds of cucumber
- Characterization of subunits encoded by SnRK1 and dissection of combinations among these subunits in sorghum (Sorghum bicolor L.)
- The role of time preferences in contract breach: Evidence from Chinese poultry farmers participating in contract farming
- Optimal design of culling compensation policy under the African swine fever — Based on simulations of typical pig farms in China
- Drip fertigation and plant hedgerows significantly reduce nitrogen and phosphorus losses and maintain high fruit yields in intensive orchards
