Identification,evolution,expression and protein interaction analysis of genes encoding B-box zinc-finger proteins in maize
2023-02-03XUXiaohuiLlWenlanYANGShukeZHUXiangzhenSUNHongweiLlFanLUXingboCUlJinjie
XU Xiao-hui ,Ll Wen-lan ,YANG Shu-ke ,ZHU Xiang-zhen ,SUN Hong-wei ,Ll Fan ,LU Xing-bo,CUl Jin-jie
1 Shandong Key Laboratory of Plant Virology/Institute of Plant Protection,Shandong Academy of Agricultural Sciences,Jinan 250100,P.R.China
2 State Key Laboratory of Cotton Biology/Institute of Cotton Research,Chinese Academy of Agricultural Sciences,Anyang 455000,P.R.China
3 Maize Research Institute,Shandong Academy of Agricultural Sciences,Jinan 250100,P.R.China
Abstract The B-box (BBX) family of proteins consists of zinc-finger transcription factors with one or two highly conserved B-box motifs at their N-termini.BBX proteins play crucial roles in various aspects of plant growth and development,including seedling photomorphogenesis,shade avoidance,flowering time,and biotic and abiotic stress responses.Previous studies have identified many different BBXs from several plant species,although the BBX family members in maize are largely unknown.Genome-wide identification and comprehensive analysis of maize BBX (ZmBBX) expression and interaction networks would therefore provide valuable information for understanding their functions.In this study,36 maize BBXs in three major clades were identified.The ZmBBXs within a given clade were found to share similar domains,motifs,and genomic structures.Gene duplication analyses revealed that the expansion of BBX proteins in maize has mainly occurred by segmental duplication.The expression levels of ZmBBXs were analyzed in various organs and tissues,and under different abiotic stress conditions.Protein-protein interaction networks of ZmBBXs were established using bioinformatic tools and verified by bimolecular fluorescence complementation (BiFC) assays.Our findings can facilitate a greater understanding of the complexity of the ZmBBX family and provide novel clues for unravelling ZmBBX protein functions.
Keywords: maize,B-box family protein,evolution,expression,protein interaction
1.lntroduction
The B-box (BBX) family of proteins is a subgroup of the zinc-finger transcription factors that have one or two highly conserved B-box motifs with specialized tertiary structures at their N-termini (Khannaet al.2009).The B-box domain is stabilized by the binding of Zn2+ions(Klug and Schwabe 1995),and it is crucial for proteinprotein interactions and transcriptional regulation (Putterillet al.1995;Borden 1998;Torok and Etkin 2000;Robsonet al.2001).In addition to the B-box domains,some BBX proteins in plants have a CCT domain that plays important roles in transcriptional regulation and nuclear protein transport (Laubingeret al.2006;Janget al.2008;Khannaet al.2009).Furthermore,the valineproline (VP) motif,which shows high similarity to the CCT domain,appears in some BBX proteins.The VP motif has been reported to play important roles in the interactions between BBXs and COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) (Holmet al.2001;Dattaet al.2007;Lauet al.2019).
In recent years,the roles of BBXs have been elucidated,mainly in the model plantArabidopsiswhich contains 32 BBX members in its genome (Khannaet al.2009).Multiple BBX family proteins play important roles in seedling photomorphogenesis,including seedling de-etiolation,cotyledon unfolding,lateral root growth,chlorophyll accumulation,and anthocyanin production.During seedling photomorphogenesis,different BBX members have opposing functions.Some BBX proteins,such as AtBBX4 and AtBBX20-22,promote photomorphogenesis,whereas others,including AtBBX18,AtBBX19,and AtBBX32,inhibit this process (Dattaet al.2006,2007,2008;Parket al.2011;Wanget al.2011,2014,2015;Fanet al.2012).CO (CONSTANS) was the first BBX member found to have a function in flowering,and other BBXs,such as AtBBX4,AtBBX6,AtBBX7,and AtBBX32,were subsequently found to play essential roles in the control of flowering (Cheng and Wang 2005;Hassidimet al.2009;Tripathiet al.2017).In addition to plant growth and development,BBXs are involved in biotic and abiotic stress responses.AtBBX24 positively regulates salt tolerance inArabidopsis(Nagaoka and Takano 2003),while AtBBX18 negatively regulates thermotolerance(Wanget al.2013).Ectopic expression of MdBBX10 fromMalus domesticaenhances the salt and drought tolerance ofArabidopsis(Liuet al.2019).In rice,the CO-like B-box geneOsCOL9positively regulates blast resistance,and knock-out ofOsCOL9increases the possibility ofMagnaporthe oryzaeinfection (Liu Het al.2016).In recent years,the involvement ofBBXgenes in plant hormone signaling pathways has been discovered.For example,AtBBX20,AtBBX18,and AtBBX16 respectively participate in brassinolide (BR) (Wanget al.2011),gibberellic acid (GA) (Sunet al.2010),and auxin(Zhanget al.2014) signaling.
The BBX family in rice consists of 30 proteins (Huanget al.2012).These proteins have been characterized,and their whole life-cycle expression profiles and diurnal expression patterns have been described.In maize,12 double B-box zinc finger genes (DBBs) lacking a CCT domain have been implicated in light regulation (Liet al.2017).Nevertheless,our understanding of theBBXgene family in maize is still limited.
In this study,36BBXswere identified from the maize genome,and their structures,expression characteristics,phylogenetic relationships,and interactions with proteins within and outside theZmBBXfamily were comprehensively analyzed.To our knowledge,this is the first report that focuses on the entire family ofBBXgenes in maize.Because the functions ofZmBBXsare still unclear,our results provide valuable clues for understanding the roles ofBBXsin maize.
2.Materials and methods
2.1.ldentification of ZmBBXs in maize
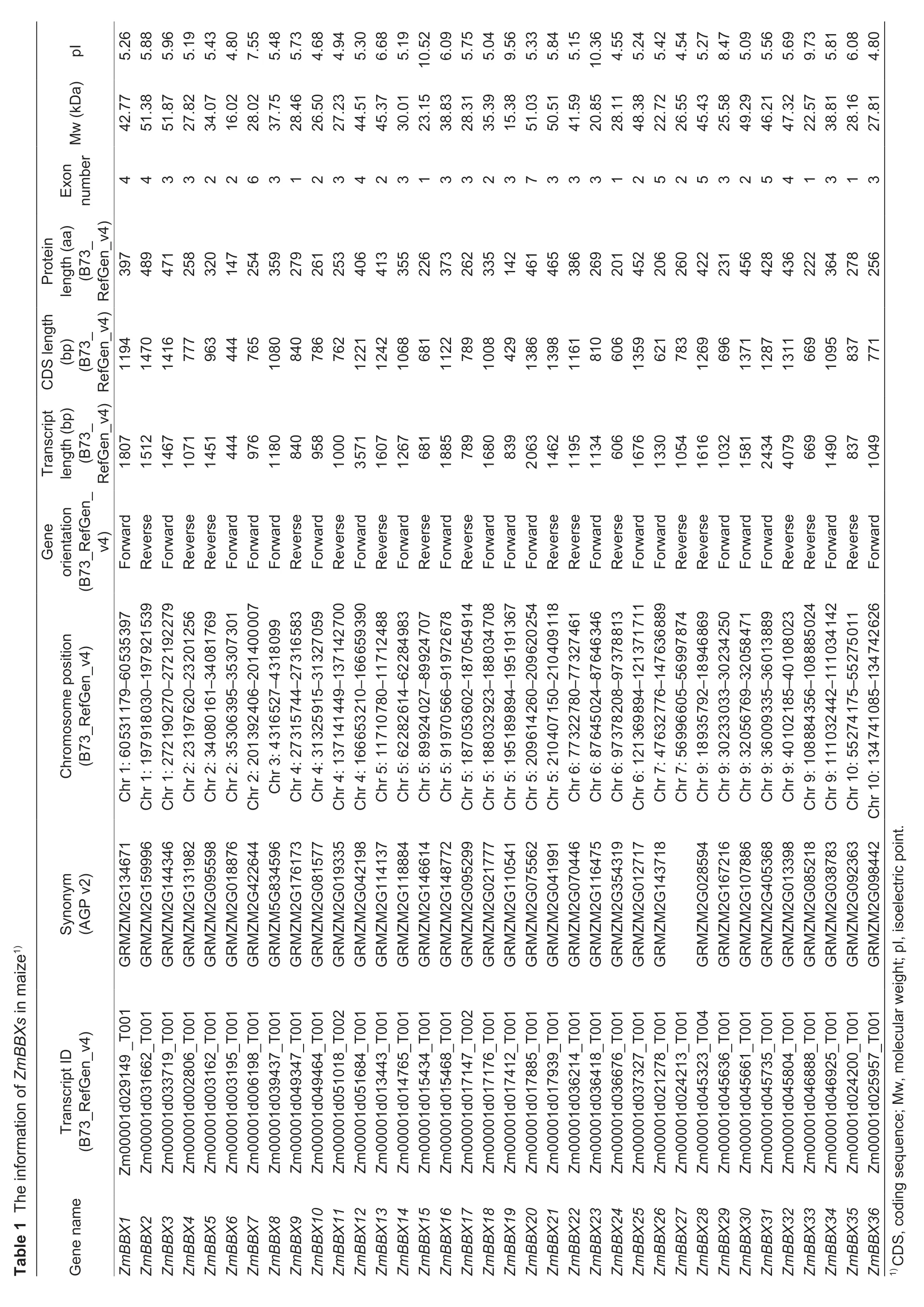
To identify candidate genes encodingZmBBXsin the maize genome,published BBX proteins fromArabidopsisand rice were used as queries in a BlastP search against the Maize Genome Database (MaizeGDB;B73 RefGen_v4,http://www.maizegdb.org/).A total of 65 protein sequences were obtained.After manually removing repetitious proteins,we identified 36 candidate BBXs.The resulting candidate ZmBBXs were then analyzed for the presence of B-box domains using the Pfam (http://pfam.xfam.org/),SMART (http://smart.embl.de/),and Prosite (https://prosite.expasy.org/) websites.Proteins containing at least one B-box domain were considered to be members of the maize BBX family.The basic characteristics of eachZmBBX,including chromosomal position,full-length cDNA and protein sequences,and gene structure,were obtained from MaizeGDB.Molecular weights and isoelectric points were calculated with ExPASY (http://web.expasy.org/compute_pi).
2.2.Chromosomal locations and gene duplication
The chromosomal location of each ZmBBX was plotted using the Map Gene2Chromosome v2 website (http://mg2c.iask.in/mg2c_v2.0/) and the information was downloaded from MaizeGDB.Gene duplication events of maizeBBXfamily members were determined according to the criteria described by Guet al.(2002) and Yanget al.(2008).
2.3.ldentification of conserved motifs
Conserved motifs in the full-length protein sequences of ZmBBXs were predicted using the MEME suite (http://meme.nbcr.net/meme) with the following parameters:optimum motif width=6-50 and the maximum number of motifs=10.Default settings were used for all other parameters.
2.4.Expression patterns of ZmBBXs obtained from the published literature
Expression data for theZmBBXsin 21 distinct tissues and organs were downloaded from a previous study (Walleyet al.2016).An expression heat map was constructed using Cluster 3.0 Software.
2.5.Phylogenetic analyses
Full-length protein sequences of ZmBBXs were aligned in ClustalW with default parameters.To determine the evolutionary relationships of ZmBBXs,a phylogenetic tree was constructed by the neighbor-joining method in MEGA v5.05.The parameters used for the phylogenetic analysis were as follows: Poisson Model,pairwise deletion,and 1 000 bootstrap replicates.
2.6.Plant materials and stress treatments
Maize seedlings were cultured as described by Xuet al.(2012).The second leaves and primary roots from 2-wk-old seedlings (vegetative stage 2 (V2 stage)) were collected as young leaf and root tissues,respectively.The mature leaves and internodes at the V14 stage were collected as leaf and internode tissues,respectively.The silks and pollen grains were harvested at the reproductive stage 1(R1 stage).The embryos and endosperms were collected from immature ears at 12-14 days after pollination (DAP).For light duration treatments,the second leaves of 2-wkold seedlings cultured under either long-day conditions (15-h light/9-h dark) or short-day conditions (12-h light/12-h dark) were collected every 4 h.For salt and drought stress treatments,2-wk-old seedlings grown under longday conditions were respectively exposed to 250 mmol L-1NaCl or 15% PEG6000 dissolved in Hoagland’s solution.After 1,4 and 8 h of the treatment,the second leaves were collected.
2.7.Cloning of the full length ZmBBX genes
The full-length coding sequences ofZmBBXswere amplified by PCR using cDNA from the leaves of 2-wk-old seedlings as the template.The amplicons of the expected sizes were cloned into the 5 min TA/Blunt-Zero Cloning vector (Vazyme,Nanjing,China) and then the positive clones were sequenced.The sequencing files were compared with the corresponding published sequence in the MaizeGDB database by DNAMAN7.0.The primers used in this experiment are listed in Appendix A.
2.8.Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted using a TransZol Up Plus RNA kit (TransGen,Beijing,China) according to the manufacturer’s instructions.After removal of genomic DNA,first-strand cDNA was synthesized using an oligo(dT) primer and EasyScript One-Step gDNA Removal and cDNA Synthesis Supermix (TransGen).qRT-PCR amplification was performed with gene-specific primers(Appendix B),appropriately diluted cDNA,and SYBR Green Master Mix (Roche Diagnostics,Mannheim,Germany).The relative expression level of each gene was calculated according to the delta-delta thresholdcycle relative quantification method,with 18S rRNA used as an internal control.For each gene,three biological replicates were carried out.
2.9.ldentification of ZmBBX-interacting proteins
Each full-length ZmBBX protein sequence was searched against the STRING database to identify known and predicted interactions,with the minimum required interaction score set to medium confidence (0.400).To construct the interaction networks between ZmBBXs,all ZmBBX protein sequences were used as queries when searching for known and predicted interactions.
2.10.Bimolecular fluorescence complementation(BiFC)
The vector pSm35s was used for the bimolecular fluorescence complementation (BiFC) assay.The fulllength coding sequences of five genes -ZmBBX4,ZmBBX14,ZmBBX19,ZmBBX23andZmBBX36-without the stop codon were amplified using the primers listed in Appendix C.HY5andbZIP6were obtained by full gene synthesis.Using the homologous recombination method,HY5andbZIP6were inserted into the pSm35scYFP vector and theZmBBXswere inserted into the pSm35s-nYFP vector provided by BioRun Company(Wuhan,China).Chloroplast auto-fluorescence was used as the control.The maize protoplast transformation was adopted as described previously (Narusakaet al.2008).YFP fluorescence in maize protoplasts was imaged at 16-24 hours after protoplast transformation using a Nikon C2-ER confocal laser scanning microscope.
3.Results
3.1.ldentification of BBX genes in the maize genome
To identify putativeBBXgenes in maize,we searched the maize genome database MaizeGDB using the amino acid sequences ofBBXgenes inArabidopsisand rice as queries.After manually removing repetitive proteins,we identified 36 candidate BBXs,including 12 previously reported DBB proteins (Liet al.2017).To confirm the presence of the B-box domain in each sequence,all protein sequences were analyzed using the Pfam,SMART,and Prosite websites.All of the 36 candidate BBXs were found to possess either one or two B-box domains,and some contained a conserved CCT domain(Fig.1;Table 1).To confirm the accuracy of theZmBBXgenes documented in the MaizeGDB,we cloned the full length of 20ZmBBXgenes.As expected,most of the sequences (17/20) amplified from the B73 tissues were the same as their corresponding sequences downloaded from MaizeGDB.However,the other threeZmBBXs(ZmBBX3,ZmBBX6andZmBBX36) showed either one or three base mutations (Appendix D).Overall,these results indicate that the sequencing project of the maize B73 inbred line is relatively accurate,but minor errors still exist in the maize genome database.The sequence data in MaizeGDB is valuable since it allows researchers to explore the functions of genes in the maize genome.The identified proteins were named asZmBBX1toZmBBX36according to their sequential locations on maize chromosomes 1-10 (Fig.2).The characteristics of these ZmBBXs,including chromosomal locus,gene orientation,length of transcript,CDS and protein,number of exons,molecular weight,and isoelectric point,are listed in Table 1.The gene structures are illustrated in Appendix E.
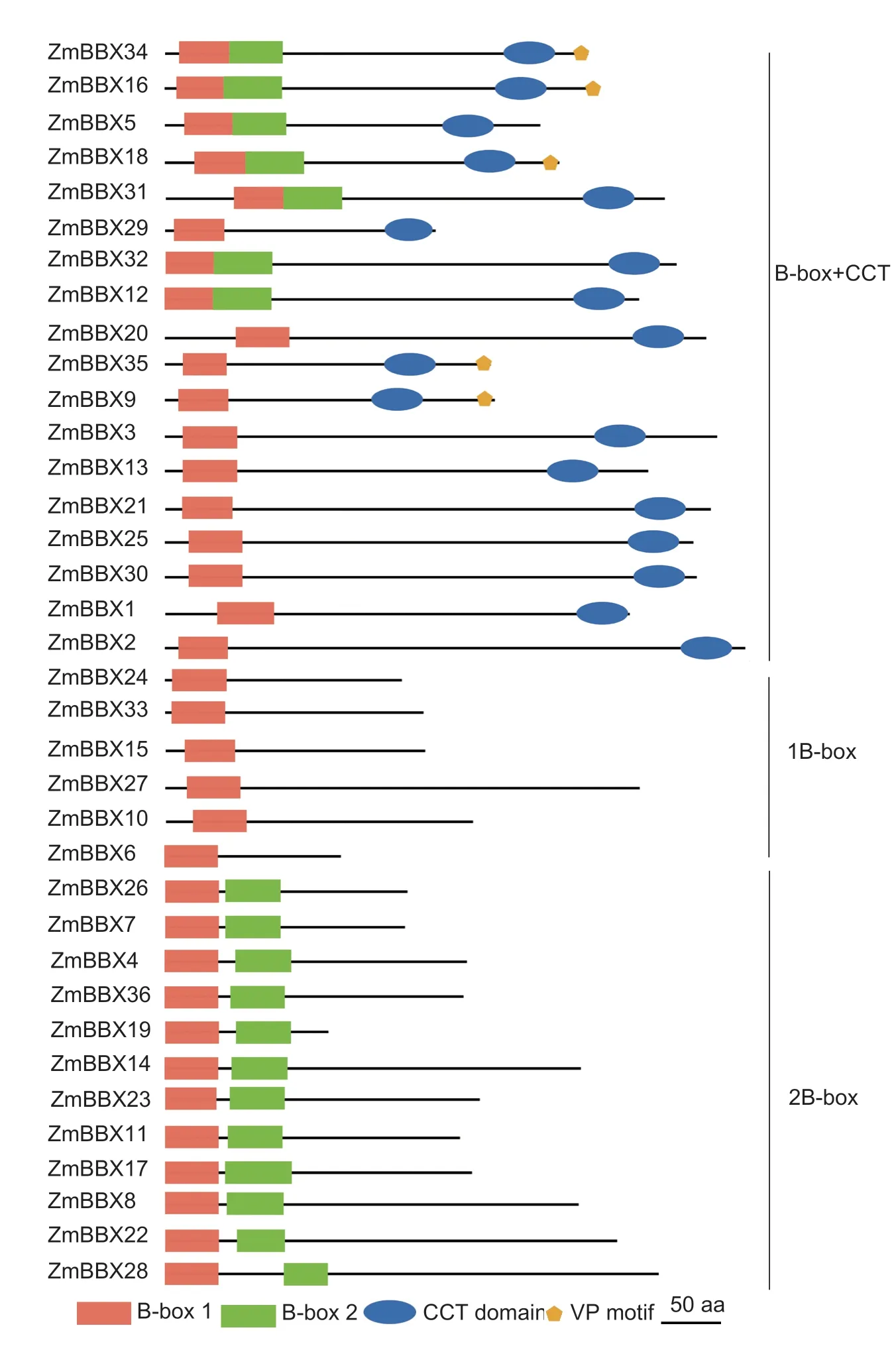
Fig.1 Protein structures of ZmBBXs.Known motifs of ZmBBXs are represented as different colored shapes.The lengths and distributions of these domains correspond to their actual sizes and locations in ZmBBX proteins.
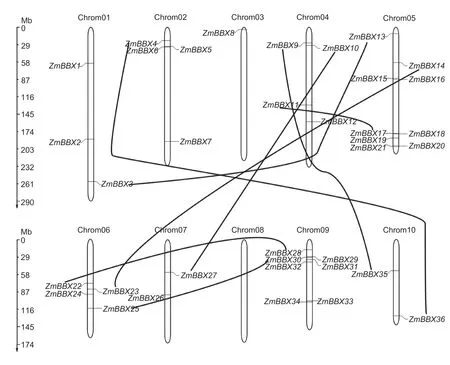
Fig.2 Chromosomal location of the ZmBBX family.Each ZmBBX gene is indicated on its corresponding chromosome.Segmentally duplicated ZmBBXs are connected by black curves.Chromosome size scales are shown on the left.Chrom,chromosome.
3.2.Phylogenetic analysis of BBX proteins in maize
To examine the phylogenetic relationships of the BBX proteins,the 36 ZmBBX protein sequences were aligned and used to construct an unrooted phylogenetic tree.As shown in Fig.3,the ZmBBX proteins were divided into three clades (I,II,and III).Clade I,which included BBX proteins with one or two B-box domains and a conserved CCT domain,could be further divided into three subclades.Subclades IB and IC consisted of BBX proteins with one B-box domain and a CCT domain,while subclade IA mainly consisted of BBX proteins with two B-box domains and a CCT domain (Fig.3).Clade II comprised five BBX proteins having only one B-box domain (Fig.3).Clade III contained 12 previously reported DBBs (Liet al.2017) plus ZmBBX7,which contained only one B-box domain.The topology of the phylogenetic tree did not strictly correspond to the distribution of B-box and CCT domains,consistent with the results of a phylogenetic analysis of OsBBXs (Huanget al.2012) but not that of AtBBXs (Khannaet al.2009).These results indicate that BBXs evolved independently after the divergence of monocots and dicots.

Fig.3 Phylogenetic tree of ZmBBX family members.Neighbor-joining phylogenetic tree constructed in MEGA v5.05 from fulllength amino acid sequences of ZmBBXs aligned in ClustalW.Bootstrap support values (%) based on 1 000 replicates are shown at each node.The scale bar represents 0.1 amino acid substitutions per site.The suffixes B,BC,and BBC following the names of ZmBBXs in the phylogenetic tree indicate which domains they contain: B,only a B-box domain;BB,two B-box domains;BC,a B-box and a CCT domain;BBC,two B-box domains and a CCT domain.
We also generated a phylogenetic tree of the BBXs fromArabidopsis,rice,and maize (Appendix F).The BBXs from these three plant species were divided into five groups.Groups I-III comprised BBX proteins that possess one or two B-box domains with CCT domains,whereas groups IV-V contained BBXs but no CCT domain.The clade placement of most ZmBBXs in this tree was the same as that in the phylogenetic tree constructed using only the ZmBBXs (Appendix F).Within each clade,paralogous BBXs from rice and maize clustered together and were separate from most of the BBX orthologs fromArabidopsis(Appendix F).
To investigate the events underlyingZmBBXgene family evolution,we calculated the physical distances,coverage,and amino acid similarities.No gene clusters were found in theZmBBXgene family,but eight segmental duplication events were identified between the chromosomes (ZmBBX3and13,ZmBBX4and36,ZmBBX 9and35,ZmBBX10and27,ZmBBX11and17,ZmBBX14and23,ZmBBX22and28,andZmBBX25and30) (Fig.2).These results suggest that theZmBBXgene family expanded through segmental duplication,not tandem amplification.
3.3.Domains and motifs of the BBX proteins in maize
The distributions of the three domains among the maize BBX proteins are shown in Fig.1.To identify more conserved motifs,we analyzed all of the amino acid sequences of ZmBBXs using the MEME server (http://meme-suite.org/tools/meme).As expected,the top three conserved motifs identified by MEME overlapped with the three characteristic domains of BBX proteins,i.e.,motifs 1-3 were included in the B-box 1,CCT,and B-box 2 domains,respectively (Fig.4;Appendix G).Motifs 4,6,and 7 were included in the CCT,B-box 1,and B-box 2 domains,respectively (Fig.4;Appendix G).The novel motif 9 was found only in group IB ZmBBXs,while motif 5 only occurred in half of the group III ZmBBXs.New motifs 5 and 9 may be useful for the identification of BBX proteins in other plant species.

Fig.4 Top 10 conserved motifs in ZmBBXs identified by MEME analysis.
3.4.Expression patterns of ZmBBXs in various tissues and organs
The expression profiles of genes are generally related to their functions.To investigate the expression patterns ofBBXgenes during maize development,information on the global transcription profiles in 21 distinct maize tissues were collected from the published literature(Walleyet al.2016).As shown in Fig.5,all 36ZmBBXswere expressed in at least one examined tissue.SomeBBXswere preferentially expressed in specific tissues,includingZmBBX9andZmBBX29in mature pollen,ZmBBX25andZmBBX30in mature leaf tissues,andZmBBX15in embryos at 20 DAP.Conversely,ZmBBX28was constitutively expressed in almost all of the examined tissues.Interestingly,the ZmBBXs with similar protein structures,such asZmBBX3/ZmBBX13andZmBBX7/ZmBBX26,had similar expression patterns.

Fig.5 Expression profile of ZmBBX genes in different maize tissues and organs.Heatmap based on log2-transformed expression data.The relative expression level of each gene in a tissue or organ is represented according to the color scale in the upper left-hand corner of the map,with yellow and red corresponding to lower and higher levels of expression,respectively.Gray indicates that no expression was detected.DAP,days after pollination.
In order to validate the expression patterns ofZmBBXgenes in different tissues and organs obtained from the published literature (Walleyet al.2016),qRTPCR analysis of 24ZmBBXgenes (excluding the 12ZmDBBgenes) was performed on eight different tissues(Fig.6).As expected,20 of the genes showed highly similar expression profiles between the qRT-PCR data and the published literature data (Figs.5 and 6).For example,ZmBBX15,ZmBBX25andZmBBX27exhibited higher expression levels in silks,which corresponded with the data from the published database.However,theZmBBX2,ZmBBX9,ZmBBX12andZmBBX24genes showed some differential expression patterns relative to the published data.ZmBBX2was highly expressed in the embryo in the published data,whereas the qRT-PCR result showed higher expression in silks.The expression ofZmBBX9was higher in mature leaf than in internode,the expression ofZmBBX12was higher in mature leaf and silks than in primary root,andZmBBX24was higher in silks than in the embryo.The conflicting results between qRT-PCR and microarray data may be due to the differences in plant growth conditions,experimental conditions and data processing methods.

Fig.6 Expression patterns of the ZmBBX genes in different tissues by qRT-PCR.The 18S rRNA gene was used as internal control in the qRT-PCR.Bars mean SD (n=3).

Fig.6 (Continued from preceding page)
3.5.Diurnal expression patterns of ZmBBXs
ManyBBXgenes in Arabidopsis and rice have diurnal expression patterns (Huanget al.2012).To investigate the diurnal expression patterns of theBBXgenes in maize,14ZmBBXsexpressed in leaf tissues were selected for validation by qRT-PCR.The analysis revealed that 12 of the 14ZmBBXshad diurnal expression patterns,and onlyZmBBX17andZmBBX18exhibited no obvious diurnal expression pattern under the two tested photoperiodic conditions (Appendix H).Some of theZmBBXsexhibited similar diurnal expression patterns.For example,the expression ofZmBBX9andZmBBX10began to increase early in the morning,reached a peak around midday,then immediately dropped in the afternoon,and the lowest expression occurred in the dark under both photoperiodic conditions (Appendix H).
3.6.Responses of ZmBBXs to drought and salt stresses
Although BBXs have been widely reported to play important roles in light signaling,their roles in biotic and abiotic stress responses are poorly understood.Recent studies have revealed thatBBXgenes play major roles in stress responses (Minet al.2015;Liu Jet al.2016;Qinet al.2018).To investigateZmBBXgene responses to drought and salt stresses,we used qRT-PCR to examine the expression patterns of 15ZmBBXsunder drought and salt stress treatments lasting up to 8 h.During the first hour of drought treatment,all members were up-regulated.The expression levels of nineZmBBXsreached their peaks at 1 h of treatment and then sharply decreased (Fig.7).In contrast,the expression levels of the other sixZmBBXgenes reached their peaks at 8 h of drought treatment (Fig.7).Under salt treatment,all 15ZmBBXswere up-regulated (Fig.7).The expression levels of eightZmBBXsreached their peaks at 1 h of treatment,while peak expression levels of the other sevenZmBBXgenes were observed at 8 h of treatment.Interestingly,12ZmBBXshad similar responses to both drought and salt treatments (Fig.7),which implies that cross-talk may exist between the drought and salt signaltransduction pathways.
3.7.The protei-protein interaction network of ZmBBXs
Previous studies have revealed that interaction with other proteins is one of the major ways that the BBX proteins exert their functions.Benefitting from the development of bioinformatics,interactions between proteins can be predicted with online tools.Using the STRING database,we were able to predict interactions between ZmBBXs and proteins outside the BBX family.As shown in Fig.8,24 ZmBBXs were predicted to have interactions with other ZmBBX family proteins.Among them,three ZmBBXs(ZmBBX15,ZmBBX24,and ZmBBX33) were predicted to interact with most of the ZmBBX family members,whereas six ZmBBXs had only one interacting ZmBBX.In addition,14 ZmBBX proteins,mostly distributed in clades II and III,were found to interact with other proteins—mainly transcription factors (such as HY5) and the ubiquitin ligase protein COP1 (Appendix H).
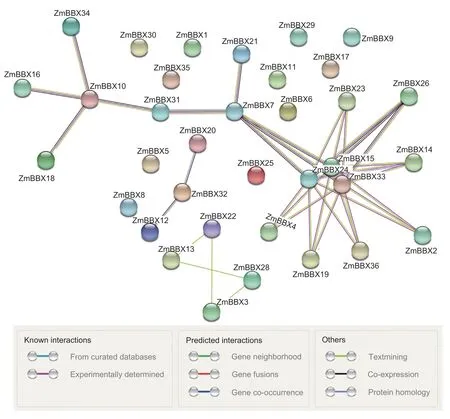
Fig.8 Network of interactions between ZmBBXs.Network nodes represent proteins,and edges represent protein-protein associations.The color of each line indicates the evidence used as the basis for the predicted interaction.The minimum required interaction score was set to medium confidence (0.400).
Next,we used bimolecular fluorescence complementation (BiFC) to validate the protein-protein interactions between ZmBBX proteins and their interacting proteins.ZmBBX4,ZmBBX14,ZmBBX19,ZmBBX23 and ZmBBX36 were fused to the N-terminal fragment of yellow fluorescence protein (YFPN),and HY5 and bZIP6 were fused to the C terminal of YFP (YFPC),to produce ZmBBX4-YFPN,ZmBBX14-YFPN,ZmBBX19-YFPN,ZmBBX23-YFPN,ZmBBX36-YFPN,HY5-YFPCand bZIP6-YFPCconstructs.As shown in Fig.9,the YFP fluorescence could be observed in the nuclei of ZmBBX4-YFPN/HY5-YFPC,ZmBBX14-YFPN/HY5-YFPC,ZmBBX19-YFPN/HY5-YFPC,ZmBBX23-YFPN/HY5-YFPC,ZmBBX36-YFPN/HY5-YFPC,ZmBBX4-YFPN/bZIP6-YFPC,ZmBBX14-YFPN/bZIP6-YFPC,ZmBBX23-YFPN/bZIP6-YFPC,and ZmBBX36-YFPN/bZIP6-YFPCcotransformed maize protoplast cells at 16-24 hours after protoplast transformation.These results indicate that various ZmBBXs can interact with HY5 and bZIP6,which is consistent with the predictions (Appendix I).
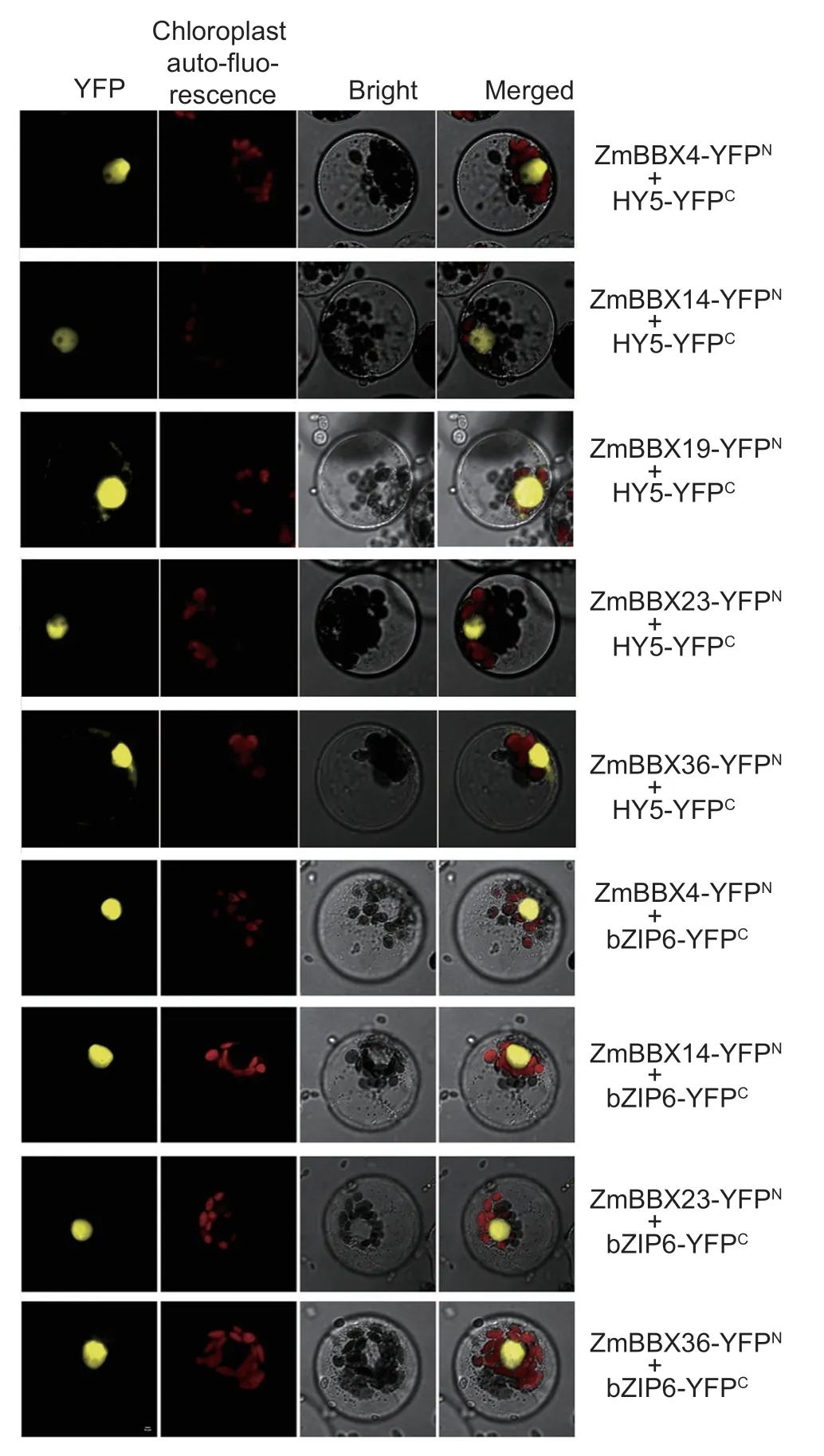
Fig.9 ZmBBXs interact with HY5 in maize protoplasts.BiFC of interactions between ZmBBX4-YFPN and HY5-YFPC,ZmBBX14-YFPN and HY5-YFPC,ZmBBX19-YFPN and HY5-YFPC,ZmBBX23-YFPN and HY5-YFPC,ZmBBX36-YFPN and HY5-YFPC,ZmBBX4-YFPN and bZIP6-YFPC,ZmBBX14-YFPN and bZIP6-YFPC,ZmBBX23-YFPN and bZIP6-YFPC,or ZmBBX36-YFPN and bZIP6-YFPC in maize protoplasts.Different vectors were co-transfected into maize protoplast cells,and the fluorescence was captured at 16-24 h after protoplast transformation using a Nikon C2-ER confocal laser scanning microscope.Bars=10 μm.
4.Discussion
4.1.Characteristics and evolution of BBX proteins in maize
BBX proteins are a class of highly conserved zinc-finger proteins that are widely distributed in both prokaryotes and eukaryotes (Zobellet al.2005;Khannaet al.2009;Yamawakiet al.2011;Croccoet al.2013).Although most BBX families in green algae consist of BBX proteins with only one B-box motif,BBXs with two B-box domains have been identified in the unicellular green algaChlamydomonas.These discoveries indicate that the BBX protein family has an ancient origin,and that the B-box duplication event occurred much earlier than the colonization of land by plants (Kenrick and Crane 1997;Peers and Niyogi 2008;Crocco and Botto 2013).
Unlike BBX proteins in animals,which all contain B-box domains,the BBX proteins in plants are diverse,with their B-box domains sometimes accompanied by a CCT domain (Borden 1998;Khannaet al.2009;Huanget al.2012).Phylogenetic analyses of BBXs from 12 plant species have demonstrated that B-box domains are common and conserved,and that the two B-box domains in plants are probably derived from segmental duplication and internal deletion events (Crocco and Botto 2013).The CCT domain,which is also highly conserved across plant species,contains a nuclear localization signal that determines the nuclear location of the BBX proteins.The numbers of CCT domain-containing BBX proteins inArabidopsis(17/32),rice (17/30),and maize (18/36)are similar (Appendix F).Besides B-box and CCT domains,several BBXs contain a VP motif consisting of a six amino acid consensus sequence G-I/V-V-P-S/T-F closely positioned near the CCT domain at the C terminus (Holmet al.2001;Dattaet al.2006).The VP motif plays an important role in mediating the interaction between BBXs and COP1.In this study,we found that five CCT domain-containing ZmBBXs have a VP motif at the C terminus,whereas the ZmBBXs with only B-box domains lack this motif (Fig.1).The VP motif distribution pattern in maize is similar to that of rice but contrasts with the pattern inArabidopsis,a species that possesses VP motif BBX proteins without CCT domains (Khannaet al.2009;Huanget al.2012).The differing VP motif distributions in these three plant species indicate that the modes of action of the VP motif-containing BBX proteins differ between monocot and dicot plant species.In addition,two novel motifs have been identified in ZmBBX proteins,motif 9 in class IB ZmBBXs and motif 5 in class III ZmBBXs (Fig.4;Appendix G).The functions of these novel motifs remain to be elucidated.
4.2.Diverse functions of BBX family proteins
BBX family proteins exert their functions in two ways:transcriptional regulation and protein-protein interactions.On the one hand,BBX proteins can directly bind to the promoter regions of their target genes to regulate target expression.For instance,AtBBX21 can bind to the T/G-box motif in the promoter ofHY5(ELONGATED HYPOCOTYL 5) to enhanceHY5expression in the light(Xuet al.2016).As another example,CO can directly bind to theFT(FLOWERING LOCUS T) promoter and activate its expression (Tiwariet al.2010).On the other hand,BBXs can form heterodimers within and outside of the BBX protein family to regulate protein functions(Dattaet al.2007,2008;Qiet al.2012;Gangappaet al.2013).For example,AtBBX21 can interact with AtBBX22 to enhance the transcriptional activity of HY5 (Dattaet al.2007;Changet al.2008).In addition,AtBBX24 and AtBBX25 can form an inactive heterodimer to repress the transcriptional activity of HY5,thereby inhibiting seedling photomorphogenesis (Gangappaet al.2013).In the present study,we identified numerous ZmBBX-interacting proteins within or outside of the ZmBBX family,most of which were either transcription factors or ubiquitin ligase protein COP1 (Fig.8;Appendix I).Various BBXs with different VP motifs have distinct affinities for binding to the WD40 motif of COP1 to regulate photomorphogenesis(Lauet al.2019).AtBBX20,AtBBX21,and AtBBX22 inhibit the activity of COP1,whereas AtBBX24 and AtBBX25 can activate COP1 (Dattaet al.2006,2007;Fanet al.2012;Gangappaet al.2013).We found that 14 ZmBBXs potentially have interactions with other ZmBBXs.Among them,ZmBBX15,ZmBBX24,and ZmBBX33 were predicted to interact with more than five other ZmBBX members (Fig.8).Other partners of ZmBBXs were also identified,most of which encode either transcription factors or COP1 (Appendix I).Verification of these interactions should help to elucidate the functional mechanisms of ZmBBXs in maize growth,development,and stress responses.
BBX proteins play central roles in light-mediated physiological and developmental processes in plants.Flowering is a precisely controlled,light-dependent process in angiosperms.Under different seasonal and photoperiodic conditions,different BBX proteins exert their respective actions on their downstream factors.InArabidopsis,CO was the first B-box protein shown to act as an activator of flowering under long-day conditions(Putterillet al.1995).It binds to the promoter ofFTto activate transcription and initiate flowering (Tiwariet al.2010).Various BBX members,such as BBX5,BBX6,and BBX7,control flowering by regulatingCOand/orFTtranscriptional activity (Cheng and Wang 2005;Hassidimet al.2009;Steinbach 2019).In addition,most BBXs are under diurnal control,but their expression patterns are variable.The expression ofCOreaches a peak in the afternoon under long days (Suárez-Lópezet al.2001).BBX4,BBX6,and BBX32 have their highest expression levels in the early morning,whereas BBX5 and BBX7 transcripts are accumulated during the night but decrease during the day (Cheng and Wang 2005).According to our study,mostZmBBXsalso show diurnal expression patterns under two photoperiodic conditions.These lightregulated ZmBBXs potentially function in the precise control of flowering under diurnal conditions.
Abiotic stresses,including drought,salinity,and extreme temperatures,drastically affect plant growth and development.Various BBX members in plants play important roles in stress responses.AtBBX18negatively regulates the heat stress response by down-regulating the heat-responsive factors DGD1 (digalactosyldiacyglycerol synthase 1),HsfA2 (heat stress transcription factor A2),and HSP101 (heat shock protein 101) (Wanget al.2013).AtBBX18-RNAi transgenic plants show improved heat tolerance,whereas AtBBX18-overexpressing transgenic plants exhibit reduced heat tolerance ability.Salt tolerance protein STO (AtBBX24) positively regulates the salinity stress response,and over-expression ofSTOinArabidopsisresults in enhanced plant tolerance to salinity stress (Nagaoka and Takano 2003).Numerous BBX genes inMalus domesticacan be induced by high salinity,cold,drought,and ABA treatment (Liuet al.2018).Among them,MdBBX10enhances plant drought and salinity tolerance by improving the ability of plants to scavenge reactive oxygen species under stress (Liuet al.2019).BBX proteins are also involved in biotic stress responses.TheCONSTANS-like geneOsCOL9regulates blast resistance by interacting with OsRACK1(Activated C-Kinase 1),which in turn can be induced by exogenous salicylic acid and ethylene (Liu Het al.2016).In the present study,we examined the expression profiles of 15BBXgenes under salinity and drought stress treatments.All testedBBXscould be induced immediately after the two abiotic stress treatments,but their expression patterns differed (Fig.7).Some members had their highest expression levels after just 1 h of stress treatment,whereas others required more time to reach their peaks.The different expression patterns ofZmBBXsunder stress treatments indicate their diverse roles in stress responses.In addition,manyZmBBXmembers,such asZmBBX9,ZmBBX10,ZmBBX12,ZmBBX13,ZmBBX15,ZmBBX18,ZmBBX20,ZmBBX21,ZmBBX26,andZmBBX35,exhibited similar expression patterns under the two different abiotic stress treatments.The similar expression patterns of these ZmBBXs under different stress treatments imply that these proteins have similar functions across different abiotic stress conditions.
5.Conclusion
In summary,36BBXgenes were identified from the maize genome and classified into three phylogenetic groups.Their gene structures,protein motif distributions,duplications,and spatial-temporal expression patterns were analyzed,and their expression was found to be activated under drought and salinity stress treatments.Multiple ZmBBX-interacting proteins were identified,and their interaction networks were established.Our results provide valuable information -not only for elucidating the function ofBBXgenes in light-signaling networks,but also for understanding their roles in stress responses.
Acknowledgements
This work was financially supported by grants from the Natural Science Foundation of Shandong Province,China (ZR2018LC005 and ZR2019BC107) and the Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences,China(CXGC2022C02).We thank Liwen Bianji,Edanz Group China (www.liwenbianji.cn/ac),for editing the English text of a draft of this manuscript.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- A 314-bp SlNE insertion in the ZNF2 promoter region may act as a repressor related to regulation of fat deposition in pigs
- An optimized protocol using Steedman’s wax for high-sensitivity RNA in situ hybridization in shoot apical meristems and flower buds of cucumber
- Characterization of subunits encoded by SnRK1 and dissection of combinations among these subunits in sorghum (Sorghum bicolor L.)
- The role of time preferences in contract breach: Evidence from Chinese poultry farmers participating in contract farming
- Optimal design of culling compensation policy under the African swine fever — Based on simulations of typical pig farms in China
- Drip fertigation and plant hedgerows significantly reduce nitrogen and phosphorus losses and maintain high fruit yields in intensive orchards
