OsMas1,a novel maspardin protein gene,confers tolerance to salt and drought stresses by regulating ABA signaling in rice
2023-02-03WANGFeibingWANChenzhongNlUHaofeiQlMingyangLlGangZHANGFanHULaibaoYEYuxiuWANGZunxinPElBaoleiCHENXinhongYUANCaiyong
WANG Fei-bing ,WAN Chen-zhong ,NlU Hao-fei ,Ql Ming-yang ,Ll Gang ,ZHANG Fan ,HU Lai-bao,YE Yu-xiu,WANG Zun-xin,PEl Bao-lei,CHEN Xin-hong,YUAN Cai-yong
1 School of Life Science and Food Engineering,Huaiyin Institute of Technology,Huai’an 223003,P.R.China
2 Huaiyin Institute of Agricultural Sciences of Xuhuai Region,Huai’an 223001,P.R.China
3 Institute of Botany,Jiangsu Province and Chinese Academy of Sciences,Nanjing 210014,P.R.China
Abstract Drought and salt stresses,the major environmental abiotic stresses in agriculture worldwide,affect plant growth,crop productivity,and quality.Therefore,developing crops with higher drought and salt tolerance is highly desirable.This study reported the isolation,biological function,and molecular characterization of a novel maspardin gene,OsMas1,from rice.The OsMas1 protein was localized to the cytoplasm.The expression levels of OsMas1 were up-regulated under mannitol,PEG6000,NaCl,and abscisic acid (ABA) treatments in rice.The OsMas1 gene was introduced into the rice cultivar Zhonghua 11 (wild type,WT).OsMas1-overexpression (OsMas1-OE) plants exhibited significantly enhanced salt and drought tolerance;in contrast,OsMas1-interference (OsMas1-RNAi) plants exhibited decreased tolerance to salt and drought stresses,compared with WT.OsMas1-OE plants exhibited enhanced hypersensitivity,while OsMas1-RNAi plants showed less sensitivity to exogenous ABA treatment at both germination and post-germination stages.ABA,proline and K+ contents and superoxide dismutase (SOD),catalase (CAT),peroxidase (POD),and photosynthesis activities were significantly increased.In contrast,malonaldehyde (MDA),hydrogen peroxide (H2O2),superoxide anion radical and Na+ contents were significantly decreased in OsMas1-OE plants compared with OsMas1-RNAi and WT plants.Overexpression of OsMas1 up-regulated the genes involved in ABA signaling,proline biosynthesis,reactive oxygen species (ROS)-scavenging system,photosynthesis,and ion transport under salt and drought stresses.Our results indicate that the OsMas1 gene improves salt and drought tolerance in rice,which may serve as a candidate gene for enhancing crop resistance to abiotic stresses.
Keywords: ABA signaling,OsMas1 gene,rice,salt and drought tolerance
1.lntroduction
Environmental abiotic stresses,such as salt and drought,affect plant growth and threaten agricultural production,making it difficult to provide and satisfy the needs of a rapidly growing population worldwide (Munns and Tester 2008;Wanget al.2016;Zhuet al.2019,2020).Plant adaptation to salt and drought stresses depends on the activation of cascades of molecular networks involved in stress perception,signal transduction,and the expression of specific stress-related genes and metabolites (Huanget al.2012;Zhu 2016).Rice is one of the most important grain crops,and its production has been significantly affected by salt and drought stresses (Jooet al.2019).Rice forms a complex defense network in resisting and adapting to abiotic stresses.Understanding the abiotic stress responses of rice is important for enhancing salt and drought tolerance.
The superfamily of α/β-hydrolase fold enzymes is one of the largest known protein families and exists widely in animals,plants,and microorganisms.The superfamily contains hydrolases (acetylcholinesterase,carboxylesterase,dienelactone hydrolase,lipase,cutinase,thioesterase,serine carboxypeptidase,proline iminopeptidase,proline oligopeptidase,and epoxide hydrolase) and the enzymes involved in reflecting activation of HCN,H2O2or O2instead of H2O for the reaction mechanism (haloalkane dehalogenase,haloperoxidase,and hydroxynitrile lyase) (Bugg 2004;Jochenset al.2011;Lenfantet al.2013;Lordet al.2013;Liuet al.2014).The ESTHER database,freely availableviaa web server (http://bioweb.ensam.inra.fr/esther) and widely used,which is the database of the α/β-hydrolase fold superfamily of protein containing >30 000 manually curated proteins,is designed to explore protein functions (Lenfantet al.2012).The biological functions of α/β-hydrolase fold enzymes in various organisms vary widely and include biosynthesis,metabolism,signal transduction,and gene expression regulation (Lordet al.2013;Liuet al.2014).To date,some reports found that a few α/β-hydrolase fold enzymes (e.g.,esterase,phospholipase D,and prolyl oligopeptidase (POP5))played important roles in responses to salt,drought,and chilling injury stresses and hormonal (abscisic acid,ABA)signaling of plants (Swapnaet al.2002;Wang 2002;Honget al.2010;Tanet al.2013;Liuet al.2014).
The maspardin protein,containing maspardin (mast syndrome,spastic paraplegia,and autosomal recessive with dementia) domain,is a member of α/β hydrolase superfamily.The maspardin protein was identified in human for the first time as an intracellular binding protein of cell surface glycoprotein CD4,which might modulate the stimulatory activity of CD4 protein and was named the acidic cluster protein 33 (ACP33) (Zeitlmannet al.2001;Hanna and Blackstone 2009;Liuet al.2014).A mutation in themaspardingene is due to a shift of reading frame and premature stop codon that cause maspardin protein truncation and presumed loss of function and result in a lack of α/β-hydrolase activity.This mutation might cause the complicated form of hereditary spastic paraplegia known as Mast syndrome (Simpsonet al.2003).Hanna and Blackstone (2009) found that maspardin interacted with an aldehyde dehydrogenase superfamily member,ALDH16A1,to help clarify the cellular pathogenesis underlying Mast syndrome.There was only one report on identifying the maspardin gene at functional levels in plants (Liuet al.2014).The expression level of a maspardin gene from a salt-tolerant sweetpotato line ND98,namedIbMas,was significantly induced by NaCl stress and ABA treatment,and theIbMas-overexpressing sweetpotato (cultivar Shangshu 19)plants exhibited enhanced salt tolerance (Liuet al.2014).However,the biological function and molecular characterization of a maspardin gene response to salt and drought stresses in rice are rarely known.
Based on the cDNA-amplified fragment length polymorphism (cDNA-AFLP) library of rice cultivar Yangjing 805 under 200 mmol L-1mannitol stress,we isolated a novel ricemaspardingene namedOsMas1.This study found that the expression level ofOsMas1was induced by abiotic stresses.In order to carry out functional studies on theOsMas1gene and later on to apply it for the genetic engineering of crops,we first developed the transgenic rice overexpressing or interfering withOsMas1,respectively.Our study clearly indicated that the expression levels of ABA signaling responsive related genes were significantly up-regulated in theOsMas1overexpression rice plants under salt and drought stresses,and the overexpression rice plants had exhibited enhanced tolerance to salt and drought stresses.All these findings will be utilized for genetically modified crops with enhanced tolerance to abiotic stresses.
2.Materials and methods
2.1.Plant materials and growth conditions
Rice varieties Yangjing 805 and Zhonghua 11 were used in this study.Yangjing 805 was employed for the cloning and expression analysis of theOsMas1gene.Zhonghua 11 (wild type,WT) was used to characterize the functions ofOsMas1in responses of the transgenic plants to salt and drought stresses.Based on the cDNAAFLP library of rice cultivar Yangjing 805 under 200 mmol L-1mannitol treatment,an expressed sequence tag(EST) showing homology to knownmaspardin-like genes exhibited an increased expression level after mannitol stress.The germinated seeds of the two varieties were grown under the same growth conditions as previously described (Liet al.2017).Three-wk-old seedlings were treated with 200 mmol L-1mannitol,20% polyethylene glycol PEG6000,200 mmol L-1NaCl,and 100 μmol L-1ABA treatments.For gene expression analysis,the leaf tissues were collected over a time course as at 0,3,6,12,24,and 48 h post each treatment of mannitol,PEG6000,NaCl,and ABA,respectively.
2.2.lsolation and sequence analysis of OsMas1 and its promoter
Total RNA (TRIzol reagent;Invitrogen,USA) and genomic DNA (EasyPure Plant Genomic DNA Kit;TransGen,China) were extracted from fresh leaves of Yangjing 805 plants.The corresponding cDNA fragments and genomic DNA sequences were amplified usingOsMas1primers(Appendix A) and analyzed as described by Zhanget al.(2019).The promoter region was cloned with Universal GenomeWalker 2.0 Kit (TaKaRa,Dalian,China).Thecis-acting regulatory elements in the promoter region ofOsMas1were screened with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The homolog genes ofOsMas1were analyzed by an online BLAST at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/).For the multiple sequence alignment analysis,the amino acid sequence ofOsMas1and otherMashomologs from different plant species retrieved from NCBI were aligned using the DNAMAN Software (Lynnon Biosoft,Quebec,Canada).The phylogenetic analysis was conducted with the MEGA4 Software (http://www.megasoftware.net/).Theoretical molecular weight and isoelectronic point (pI) were calculated using the ProtParam tool (http://web.expasy.org/protparam/).The conserved domain of the OsMas1 protein was scanned by the InterProScan Program (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
2.3.Subcellular localization
The OsMas1 ORF without stop codon amplified with specific primers was ligated into pCAMBIA1301 (Appendix A).Isolation of maize protoplasts and transfection of the vectors into the protoplasts were performed according to the method of Yooet al.(2007).The recombinant vectors 35S-OsMas1-GFP and 35S-GFP (as control) were transfected into the isolated protoplasts and incubated overnight.The fluorescent signals of the expressed proteins were observed by fluorescence microscopy(LSM710,Zeiss).
2.4.Vector construction and transformation
The full-length coding region ofOsMas1was amplified from the first-strand cDNA of Yangjing 805 using genespecific primer pairs and cloned into theBamHI andSacI sites of plant binary vector pCAMBIA1301 to generateOsMas1overexpression (OE) and RNA interference(RNAi) construction under the CaMV 35S promoter.The OE and RNAi vectors also containedgus AandhptII genes driven by a CaMV 35S promoter,respectively.Then the recombinant vectors were transformed intoAgrobacteriumtumefaciens EH105 and then transformed into Zhonghua 11 by theA.tumefaciens-mediated methods (Hieiet al.1994).Rice transformants were selected based on their resistance to hygromycin (Hyg)based on previously described methods (Liet al.2017).Positively transgenic seedlings were grown in pots containing a mixture of soil,vermiculite,and humus (1:1:1,v/v) to select T2and T3seeds.The incubation and growth conditions of rice were the same as previously published methods (Liet al.2017).
2.5.DNA extraction and PCR detection
DNA was extracted from rice leaves according to the instructions of the EasyPure Plant Genomic DNA Kit(Transgen,Beijing,China).The presence of theOsMas1expression construct in positive plants was assessed by PCR analysis using specific primers (Appendix A).PCR amplifications were performed with an initial denaturation at 94°C for 3 min,followed by 35 cycles at 94°C for 30 s,55°C for 30 s,72°C for 1 min,and final extension at 72°C for 10 min.PCR products were separated by electrophoresis on a 1.0% (w/v) agarose gel.
2.6.Salt and drought tolerance assays
To evaluate the main agronomic traits of rice plants under normal conditions,we transplantedOsMas1-overexpression (OsMas1-OE),OsMas1-interference(OsMas1-RNAi),and WT plants into 1-m-deep containers containing natural paddy soil located in the greenhouse at 26/20°C (day/night) based on the methods of El-Esawi and Alayafi (2019).The containers were arranged in a completely randomized design,and the plants were watered daily by growing under normal growth conditions till harvesting.When the grains ripened,the following yield parameters were scored;plant height (cm),panicle length (cm),the number of spikelets per panicle,the total number of spikelets per hill,the number of filled grains per hill,filling rate (%),and total grain weight (g).
For salt and drought tolerance of rice plants,the seeds ofOsMas1-OE,OsMas1-RNAi,and WT rice lines were germinated on 1/2 medium under 16 h light (28°C)/8 h dark(25°C) photoperiod conditions for 1 wk and transplanted to 1/2 MS medium with no stress,200 mmol L-1NaCl,and 200 mmol L-1mannitol,respectively,according to the methods of Liet al.(2017).Plant length and fresh weight ofOsMas1-OE,OsMas1-RNAi and WT plants were measured after 1 wk.Four-wk-old seedlings ofOsMas1-OE,OsMas1-RNAi,and WT plants were grown in 9-cm diameter pots containing a mixture of soil and vermiculite (1:1,v/v) in a greenhouse.All pots were irrigated sufficiently with water for 2 wk under optimum growth conditions.Each pot was then irrigated with a 100-mL of 200 mmol L-1NaCl solution once every 2 d for 2 wk or subjected to drought stress for 1 wk by stopping irrigation and recovery with rewatering for 2 wk.Seedlings were regarded as survivals if the fresh and green young leaves emerged after the water supply.The survival rate of these plants was observed immediately.All treatments were performed in triplicate.
With respect to the salt and drought stresses assays of the isolated leaves,leaf sections of approximately 5 cm in length fromOsMas1-OE,OsMas1-RNAi,and WT plants at seedling stages were removed and immersed in MS liquid media solutions with concentrations of 200 mmol L-1NaCl and 200 mmol L-1mannitol for 96 h,respectively.Disks floated on sterile distilled water served as the experimental control.All the leaves were subsequently cultured under 16 h light (28°C)/8 h dark(25°C) photoperiod conditions.Each treatment was repeated three times,and 10 leaves were selected for each treatment.The total chlorophyll content of the rice leaves was measured according to the methods of Patraet al.(2010).
2.7.RNA isolation,cDNA synthesis,and qRT-PCR analysis
Total RNA was extracted from the rice leaves with the RNAprep Pure Kit (Tiangen Biotech,Beijing,China).The DNase-treated RNA was reverse-transcribed into firststrand cDNA with M-MLV reverse transcriptase (Tiangen Biotech,Beijing,China) according to the manufacturer’s instructions.The cDNA solution was used as a template for PCR amplification with the specific primers (Appendix A).The expression of genes related to ABA biosynthesis and signaling pathway,proline biosynthesis,reactive oxygen species (ROS) scavenging,photosynthesis,and ion transport pathways was performed by quantitative realtime PCR (qRT-PCR) according as described previously(Wanget al.2016).The riceOsActingene was used as the endogenous control for data normalization.The gene expression was quantified with the comparativeCTmethod(Schmittgen and Livak 2008).Each analysis was repeated three times.All the primers are listed in Appendix A.
2.8.ABA content and sensitivity assays
Endogenous ABA levels were performed by indirect enzyme-linked immunosorbent assay (ELISA) as described by Wanget al.(2016).ABA sensitivity at germination and post-germination stages was determined according to the methods of Liet al.(2017).The seeds ofOsMas1-OE,OsMas1-RNAi,and WT plants were germinated on 1/2 MS medium with 2.5 μmol L-1ABA,and germination rates of the treated seeds were calculated after 5 d.The seeds ofOsMas1-OE,OsMas1-RNAi,and WT plants germinated on 1/2 MS medium with no stress were transplanted to 1/2 MS medium with 2.5 μmol L-1ABA,then shoot and root lengths and fresh weight of all lines were measured after 1 wk.
2.9.Measurement of photosynthesis
Photosynthetic rate,stomatal conductance,and transpiration rate in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants grown in pots and incubated for 1 wk under optimum growth condition,for 1 wk under 200 mmol L-1NaCl stress,or for 1 wk under drought stress were measured according to the methods of Liuet al.(2014).Relative chlorophyll content (SPAD value in fresh leaves)was measured as described by Fernández-Falcónet al.(2006) with Chlorophyll Meter SPAD-502 (Minolta,Japan).The experiments were conducted from 9 to 11 a.m.on sunny days.
2.10.DAB and NBT staining
The 3,3´-diaminobenzidine (DAB) staining and nitro blue tetrazolium (NBT) staining were performed as described by Jianget al.(2016).Leaf sections of approximately 5 cm in length fromOsMas1-OE,OsMas1-RNAi,and WT plants at seedling stages grown in pots and incubated for 1 wk under optimum growth conditions,for 1 wk under 200 mmol L-1NaCl stress,or for 1 wk under drought stress were cut and soaked in a 1% solution of DAB in 50 cm Tris-HCl buffer and in a 0.1% solution of NBT in 10 cm potassium phosphate buffer for hydrogen peroxide(H2O2) and superoxide anion radicaldetection,respectively.
2.11.Measurements of the related components
2.12.Analyses of the corresponding enzyme activities
The 9-cis-epoxycarotenoid dioxygenase (NCED) activity in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants grown in pots and incubated for 1 wk under optimum growth conditions,for 1 wk under 200 mmol L-1NaCl stress,or for 1 wk under drought stress was measured with Plant NCED ELISA Kit (Uscn Life Science Inc.Shanghai,China).The activities of pyrroline-5-carboxylate synthase (P5CS),superoxide dismutase(SOD),catalase (CAT),and peroxidase (POD) above materials were measured according to the methods of Hayzer and Leisinger (1980),Wanget al.(2016),and Li Wet al.(2020),respectively.
2.13.Statistical analysis
The experiments were repeated three times.The data were presented as the mean±standard error (SE).Data differences were analyzed by Student’st-test in a twotailed analysis.Values ofP<0.05 orP<0.01 were considered to be statistically significant.
3.Results
3.1.lsolation and characterization of OsMas1
TheOsMas1gene was cloned from rice cultivar Yangjing 805 by RT-PCR.The ORF is 1 164 bp in size and encoded a polypeptide that is 387 amino acids in length.The theoretical molecular weight of the protein is 42.58 kD,and the theoretical pI is 6.45.Sequence analysisviathe InterPro Program (http://www.ebi.ac.uk/interpro/) showed that the OsMas1 protein contains a typical maspardin domain and belongs to α/β-hydrolase superfamily (Fig.1-A).The genomic DNA ofOsMas1was 5 094 bp in length.An analysis of the genomic structure using the Spidey Program revealed thatOsMas1contains eight exons and seven introns.
A BLASTP search indicated that the amino acid sequence of OsMas1 showed 57.66 to 82.95% amino acid identity with predicted protein products ofNicotiana tabacum(XP_016448831),Solanum tuberosum(XP_006363736),Ipomoea batatas(AII99394),Vitis vinifera(XP_002267811),Glycine max(XP_003525758),Brachypodium distachyon(XP_003573810),Setaria italica(XP_004983322),andZea mays(NP_001150628)(Fig.1-A).Phylogenetic analysis revealed that OsMas1 had a close relationship with the predicted protein products ofS. italicaandZ. mays(Fig.1-B).
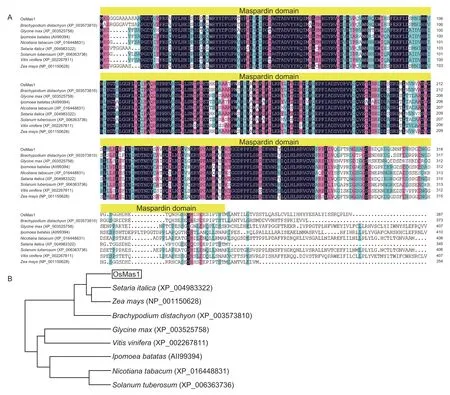
Fig.1 Analyses of the OsMas1 gene from rice.A,sequence alignment of the OsMas1 protein with its homologous proteins from other plant species.The maspardin domain is marked by yellow boxes.B,phylogenetic tree of the OsMas1 protein with its homologous proteins from other plant species.The branch lengths are proportional to distance.
A 2 109-bp fragment corresponding to the promoter ofOsMas1was isolated from Yangjing 805 genomic DNA using genome walking and was found to contain numerous types ofcis-acting regulatory elements.Among these,several kinds ofcis-acting regulatory elements involved in different abiotic stresses were identified,such as ABRE,MBS,TCA-element,CGTCA-motif,and TGACG-motif(Appendix B).The presence of these stress-relatedcisacting elements in the promoter regions indicates that the expression level ofOsMas1might be influenced by different kinds of abiotic stresses (Appendix B).
3.2.Expression of OsMas1 is up-regulated by mannitol,PEG6000,NaCl and ABA
To speculate the function ofOsMas1,we analyzed the transcript levels ofOsMas1in response to mannitol,PEG6000,NaCl,and ABA in the leaf tissues of rice cultivar Yangjing 805 by qRT-PCR.TheOsMas1transcript levels were significantly induced by 200 mmol L-1mannitol,20% PEG6000,200 mmol L-1NaCl,and 100 μmol L-1ABA,respectively (Appendix B).An increase in theOsMas1transcript was observed after 3 h of exposure to 200 mmol L-1mannitol,peaked at 6 h with a 4.57-fold higher expression level than that of untreated control,and thereafter declined (Appendix B).The expression ofOsMas1reached the highest level(5.64 folds) at 12 h of 20% PEG6000 stress,followed by a decrease (Appendix B).For 200 mmol L-1NaCl stress,the expression ofOsMas1was induced to the highest level (2.87 folds) at 6 h,followed by a decrease(Appendix B).Its expression peaked (10.57 folds) at 24 h of exposure to 100 μmol L-1ABA treatment and declined(Appendix B).These data suggest thatOsMas1was responsive to multiple abiotic stresses preferentially of rice.
3.3.OsMas1 is localized to the cytoplasm
To study the subcellular localization of OsMas1,we fused the ORF ofOsMas1with GFP and transfected it into the isolated maize protoplasts.As shown inAppendix C,the subcellular localization of maize protoplasts showed that OsMas1 was localized to the cytoplasm.
3.4.Overexpression of OsMas1 enhances salt and drought tolerance in rice
To explore whetherOsMas1plays an important role in improving agronomic traits through gene manipulation approaches,we producedOsMas1-overexpression(OsMas1-OE) plants andOsMas1-interference (OsMas1-RNAi) plants of rice cultivar Zhonghua 11 (WT).Multiple lines were obtained from Hyg resistance selection.NineOsMas1-OE rice plants,named OE1 to OE9,and fiveOsMas1-RNAi rice plants,named Ri1 to Ri5,were obtained (T1generation),and their progenies (T3generation) were generated.PCR analysis of genomic DNA indicated that they were transgenic (Appendices D and E).qRT-PCR analyses revealed significant increases in the transcript level ofOsMas1inOsMas1-OE lines,especially OE2,OE4,and OE7,and dramatic decreases inOsMas1-RNAi lines,especially Ri2 and Ri3,compared with that in WT (Appendices D and E).Therefore,OsMas1-OE lines (OE2,OE4 and OE7) andOsMas1-RNAi lines (Ri2 and Ri3) were selected for further experiments.
Under normal conditions,there were no significant differences in plant height,root length,and fresh weight betweenOsMas1-OE,OsMas1-RNAi,and WT lines grown at seedlings stages (Fig.2-A).At the tillering stages,there were no developmental differences betweenOsMas1-OE,OsMas1-RNAi and WT lines when normal irrigation (Fig.2-B).Grain yield components of different rice plants at harvesting stages were measured under normal conditions.No significant differences in plant height,panicle length,the number of spikelets per panicle,the total number of spikelets per hill,the number of filled grains per hill,filling rate,and total grain weight betweenOsMas1-OE,OsMas1-RNAi,and WT lines grown at harvesting stages under normal conditions (Appendix F).The evaluation of these yield components showed no obvious differences in the grain yield between these rice lines (Appendix F).
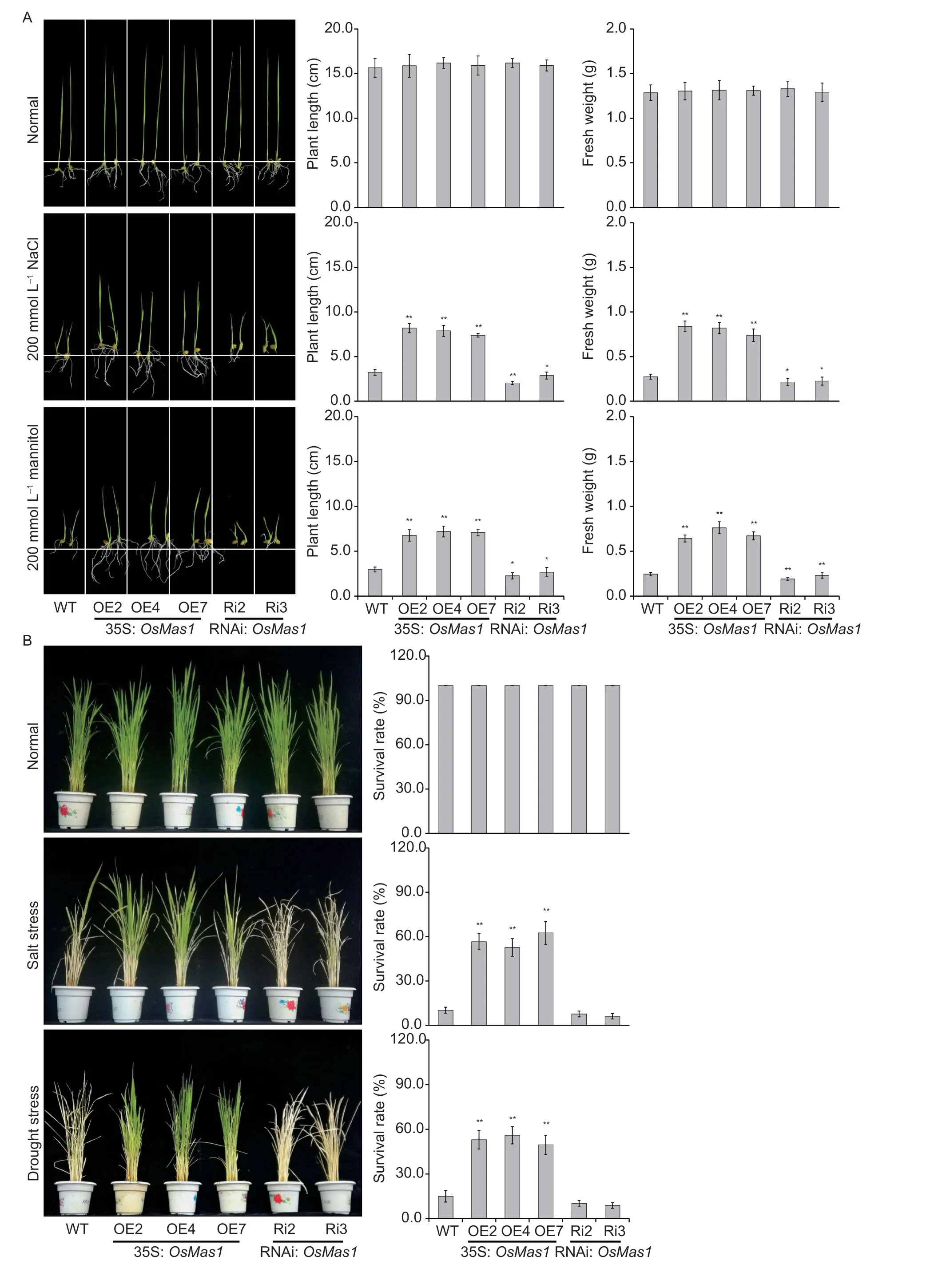
Fig.2 Enhanced salt and drought tolerance of OsMas1-OE plants.A,growth performance of OsMas1-overexpression (OsMas1-OE)rice plants,OsMas1-interference(OsMas1-RNAi) rice plants,and wild-type (WT) rice plants under 200 mmol L-1 NaCl and 200 mmol L-1 mannitol,respectively.Plant length and fresh weight of 7th-d-old seedlings after transplanting were examined.B,responses of OsMas1-OE,OsMas1-RNAi,and WT plants grown in pots and incubated for 2 wk under normal conditions,for 2 wk under 200 mmol L-1 NaCl stress,and for 1 wk by stopping irrigation and recovery with rewatering for 2 wk,respectively.Survival rates of OsMas1 overexpression,interference,and WT rice plants after salt and drought stresses were examined.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
ThreeOsMas1-OE lines (OE2,OE4,and OE7)andOsMas1-RNAi lines (Ri2 and Ri3) were selected to verify the functions of theOsMas1gene to the responses to stress treatments.The performance ofOsMas1-OE,OsMas1-RNAi,and WT lines under osmotic stress by adding 200 mmol L-1NaCl or 200 mannitol were examined,respectively.Notably,no developmental differences were identified betweenOsMas1-OE,OsMas1-RNAi,and WT lines under control growth conditions (Fig.2-A).Under salt and mannitol stresses,the growth of theOsMas1-OE seedlings was less inhibited,which exhibited higher plant length and fresh weight than these of theOsMas1-RNAi and WT seedlings (Fig.2-A).Together,these results suggest that overexpression ofOsMas1in rice could enhance the tolerance to salt and mannitol stresses at the seedling stage.
To investigate the functions ofOsMas1in response to salt and drought stresses,we grewOsMas1-OE,OsMas1-RNAi,and WT seedlings in the soil and well-watered them at the tillering stage.There were no developmental differences betweenOsMas1-OE,OsMas1-RNAi,and WT plants when normal irrigation was performed (Fig.2-B).After 2 wk of 200 mmol L-1NaCl stress,or after 1 wk of stopping irrigation and 2 wk of recovery,OsMas1-OE seedlings remained green and showed continuous growth,whereas WT andOsMas1-RNAi seedlings showed severe leaf rolling and wilting (Fig.2-B).This study also determined the survival rate forOsMas1-OE,OsMas1-RNAi,and WT seedlings grown in the soil with salt and drought stresses.As shown in Fig.2-B,the survival rates ofOsMas1-OE lines were significantly higher than that of WT andOsMas1-RNAi seedlings when exposed to salt and drought stresses.Therefore,it is evident that overexpression ofOsMas1results in enhanced tolerance to salt and drought stresses in rice.
As an explanation for the grown performance exhibited by the different transgenic plants,the possibility of a correlation between grown performance and the amount of chlorophyll retained in these plants during growth under salt and drought stresses was sought.For such experiments,uniformly cut leaf disks fromOsMas1-OE,OsMas1-RNAi,and WT plants were incubated in the presence of an indicated concentration of NaCl or mannitol for 96 h at 28°C,and the “greenness” of the plants was compared against the control set without any stress (Appendix G).Although not very well discernible visually,the actual determination of chlorophyll content in the leaf disks showed thatOsMas1-OE plants exhibited higher total chlorophyll content than WT andOsMas1-RNAi plants (Appendix G).This result indicated that theOsMas1-OE plants could retian the chlorophyll to a greater extent than WT andOsMas1-RNAi plants during growth under stress treatments.These results indicate that overexpression ofOsMas1in rice increases salt and drought tolerance.
3.5.Overexpression of OsMas1 increases sensitivity to exogenous ABA in rice
To test whether overexpression of theOsMas1gene can affect the sensitivity of transgenic rice to exogenous ABA,we germinated theOsMas1-OE and WT seeds was germinated in 1/2 MS medium with ABA (0 and 2.5 μmol L-1).As shown in Fig.3-A,the germination rate ofOsMas1-OE lines was similar to that of WT control at 0 μmol L-1ABA.However,the germination rate ofOsMas1-OE lines was significantly decreased at 2.5 μmol L-1ABA.These results suggested that the sensitivity of seed germination ofOsMas1-OE plants to ABA was increased.
Further,we tested the effect of ABA on seedling development inOsMas1-OE,OsMas1-RNAi,and WT plants.The performance ofOsMas1-OE,OsMas1-RNAi,and WT seedlings grown for 2 wk in 1/2 MS medium with or without ABA was observed (Fig.3-B and C).There were no significant differences in shoot and root lengths and fresh weight betweenOsMas1-OE,OsMas1-RNAi,and WT plants grown in MS medium without ABA (Fig.3-B and C).The shoot and root lengths and fresh weight ofOsMas1-OE plants grown in MS medium with 2.5 μmol L-1ABA were significantly reduced(Fig.3-B),while these indicators ofOsMas1-RNAi plants were clearly higher compared with those of the control (Fig.3-C).These results demonstrate that overexpression ofOsMas1makes transgenic seedlings hypersensitive to ABA in comparison to WT plants,indicating thatOsMas1may be a positive regulator of ABA signaling in rice.
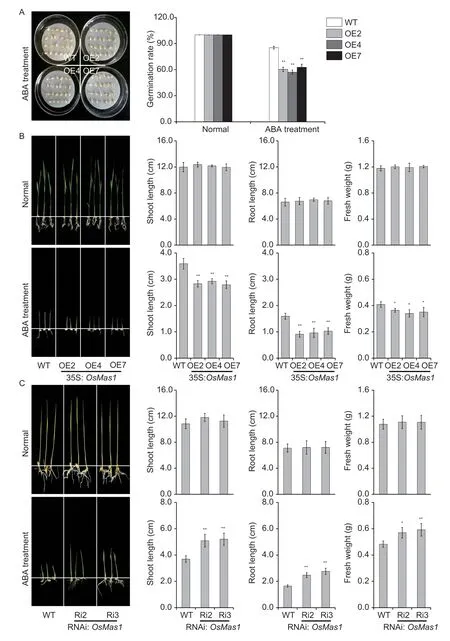
Fig.3 Increased abscisic acid (ABA) sensitivity of OsMas1-OE plants during germination and seedling stages.A,germination assay of OsMas1-OE and wild type (WT) seeds in 1/2 MS medium with 2.5 μmol L-1 ABA.The germination rates of the treated seeds were calculated after 5 d.B,growth performance of OsMas1-OE and WT plants grown in 1/2 MS medium containing 0 and 2.5 μmol L-1 ABA.C,growth performance of OsMas1-RNAi and WT plants grown in 1/2 MS medium containing 0 and 2.5 μmol L-1 ABA.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and <0.01,respectively,by Student’s t-test.
3.6.Overexpression of OsMas1 activates endogenous ABA signaling in rice
Endogenous ABA levels were measured in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses.The results showed that after salt and drought stresses,endogenous ABA levels were clearly increased inOsMas1-OE,OsMas1-RNAi,and WT lines;accumulated more ABA levels were observed inOsMas1-OE lines than that in WT andOsMas1-RNAi lines,while no obvious difference was found under normal conditions (Fig.4).It is hypothesized thatOsMas1gene may be involved in regulating salt and drought tolerance by ABA biosynthesis.
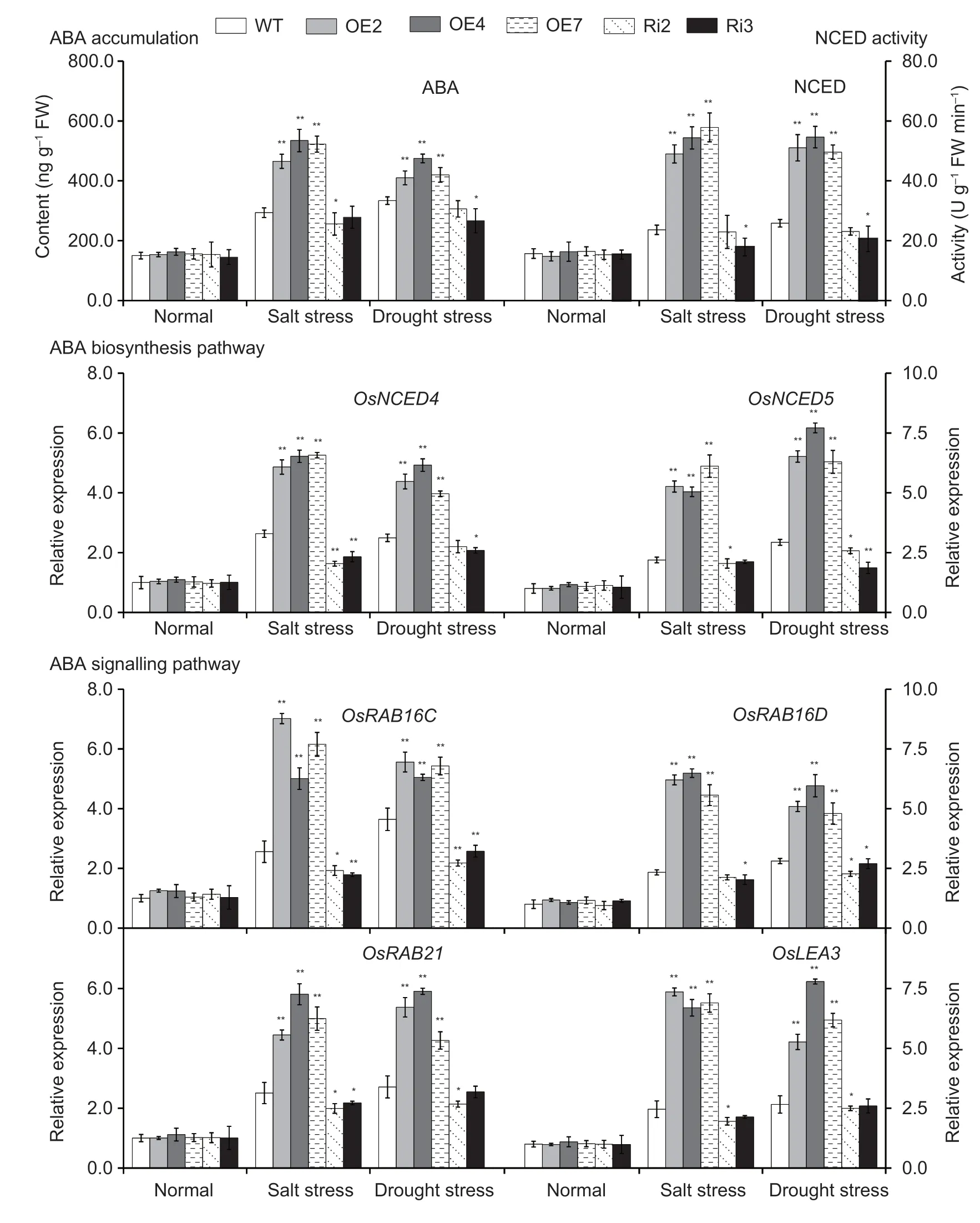
Fig.4 Abscisic acid (ABA) accumulation and 9-cis-epoxycarotenoid dioxygenase (NCED) activity and the expression of ABA biosynthesis and signaling pathways marker genes in the leaves of OsMas1-OE,OsMas1-RNAi,and wild-type (WT) plants under salt and drought stresses.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
To further confirm this hypothesis,we analyzed the activity of NCED,a key rate-limiting enzyme of ABA biosynthesis,and the transcript levels ofOsNCED4andOsNCED5inOsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses.The activity of NCED and the expression ofOsNCED4andOsNCED5were enhanced inOsMas1-OE lines compared with WT lines(Fig.4).Further research found that transcript levels of ABA-responsive signaling pathway genes,such asOsRAB16C,OsRAB16D,OsRAB21andOsLEA3,were markedly up-regulated inOsMas1-OE lines compared with WT lines under salt and drought stresses (Fig.4).TheOsMas1-RNAi lines displayed an opposite change for the respective index (Fig.4).These results indicate that theOsMas1gene may play an important role in regulating salt and drought tolerance by increasing the expression of ABA biosynthesis genes,such asOsNCED4andOsNCED5,and up-regulating the expression ofOsRAB16C,OsRAB16D,OsRAB21,andOsLEA3genes related to ABA-responsive signaling pathway.
3.7.Overexpression of OsMas1 promotes proline accumulation in rice
We analyzed the proline content,the activity of P5CS and the expression levels ofOsP5CS1andOsP5CS2,were analyzed in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses.These results showed that proline accumulation was much more significant inOsMas1-OE lines than inOsMas1-RNAi and WT seedlings under salt and drought stresses (Fig.5).P5CS activity was significantly enhanced inOsMas1-OE lines compared withOsMas1-RNAi and WT lines (Fig.5).The expression levels ofOsP5CS1andOsP5CS2,key rate-limiting enzyme genes related to proline biosynthesis,were up-regulated inOsMas1-OE lines than inOsMas1-RNAi and WT seedlings under salt and drought stresses(Fig.5).Our data indicate thatOsMas1may play a critical role in stress-induced proline biosynthesis by upregulating proline biosynthesis genes.
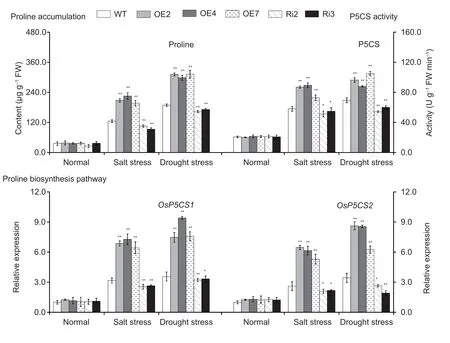
Fig.5 Free proline content and pyrroline-5-carboxylate synthase (P5CS) activity and the expression of proline biosynthesis genes in the leaves of OsMas1-OE plants,OsMas1-RNAi plants,and wild-type (WT) plants under salt and drought stresses.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
3.8.Overexpression of OsMas1 enhances photosynthesis in rice
Photosynthesis in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses was measured.The tolerantOsMas1-OE plants maintained significantly higher photosynthetic rate,stomatal conductance,transpiration rate,and chlorophyll relative content than WT,whereas theOsMas1-RNAi plants displayed an opposite change for the respective index (Fig.6).Meanwhile,we performed qRT-PCR assays to further analyze the mRNA levels of genes related to photosynthesis system under salt and drought stresses.Our data indicated that the expression levels ofOsrbcL,OspsaA,OspsbA,andOsPRKgenes,which encode Rubisco large subunit,photosystem I P700 chlorophyllaapoprotein A1,photosystem II D1 protein,and phosphoribulokinase,respectively,were up-regulated under salt and drought stresses inOsMas1-OE plants compared with WT andOsMas1-RNAi plants (Fig.6).As a consequence,it is thought that overexpression ofOsMas1enhances photosynthesis capacity,resulting in improved salt and drought tolerance in rice.
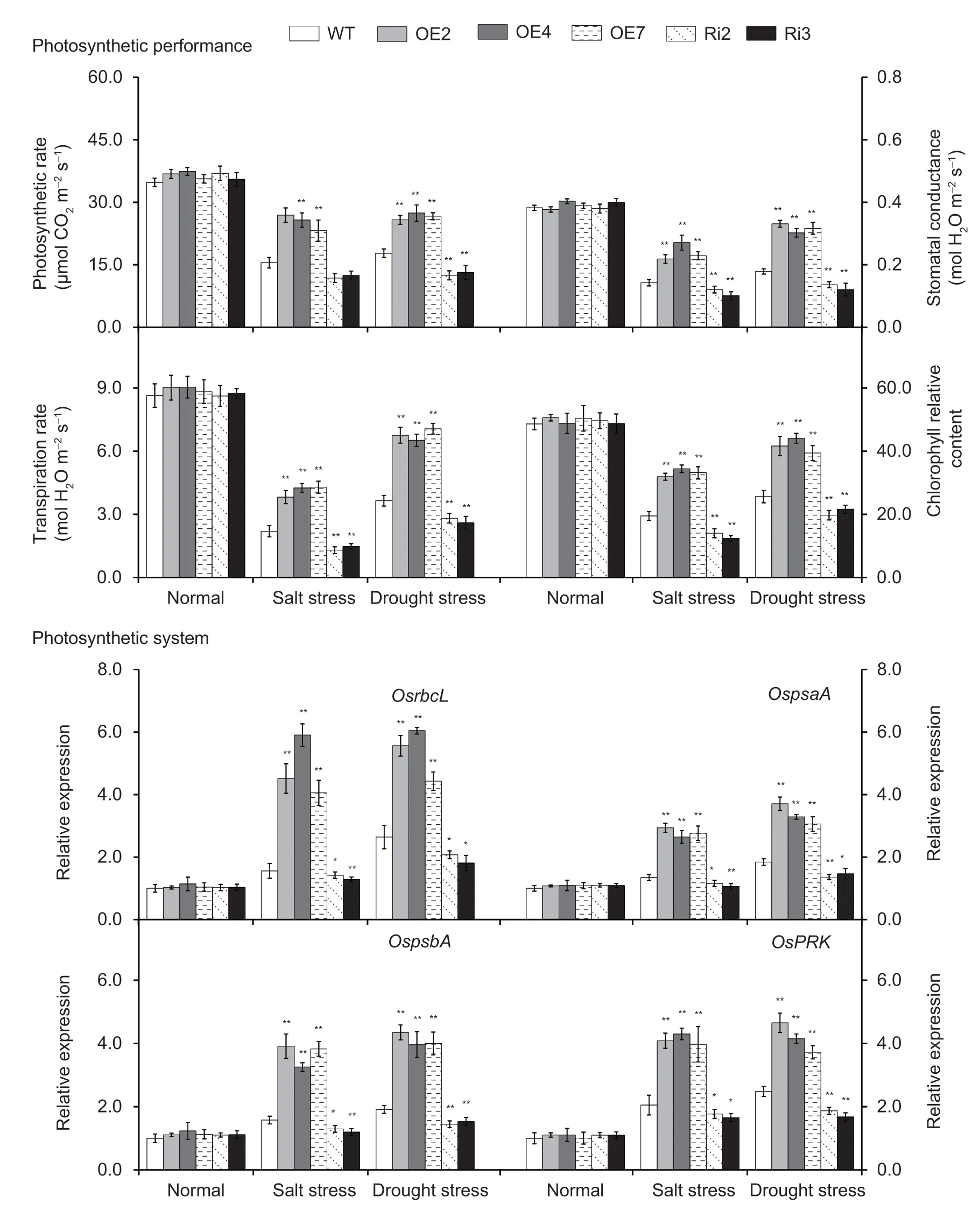
Fig.6 Photosynthetic rate,stomatal conductance,transpiration rate,and chlorophyll relative content and the expression of the genes related to photosynthesis system in the leaves of OsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
3.9.Overexpression of OsMas1 decreased ROS damage in rice
Stress usually induces damageviaoxidative damage in plants,including the generation of ROS,represented H2O2and(Jianget al.2016).The levels ofH2O2andaccumulation were evaluated by means of DAB and NBT staining and H2O2andmeasurements in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants under salt and drought stresses.Our study showed thatOsMas1-OE plants accumulated less H2O2andwhileOsMas1-RNAi plants had higher H2O2andcontents than WT plants under salt and drought stresses,whereas these were not significantly different betweenOsMas1-OE,OsMas1-RNAi,and WT plants without stress (Appendices H and I).Meanwhile,we found thatOsMas1-OE plants had significantly lower MDA than WT andOsMas1-RNAi plants (Fig.7).
Following salt and drought stresses,the activities of antioxidant enzymes,such as SOD,CAT,and POD,playing an important role in ROS-scavenging mechanisms,were measured inOsMas1-OE,OsMas1-RNAi,and WT plants.Under the absence of stress,SOD,CAT,and POD activities were not different;after salt and drought stresses,the activities of these enzymes were significantly enhanced inOsMas1-OE plants compared with those in WT andOsMas1-RNAi plants (Fig.7).qRT-PCR was used to further analyze the expression levels ofOsCu/Zn-SOD1,OsCu/Zn-SOD2,OsCatB2,andOsPOD8.1in rice plants.Under salt and drought stresses,the expression levels of these genes exhibited markedly higher expression inOsMas1-OE plants than in WT andOsMas1-RNAi plants (Fig.7).These results suggested that overexpression of theOsMas1gene may enhance the ROS-scavenging ability to decrease ROS damage,leading to improved salt and drought tolerance of transgenic rice plants.
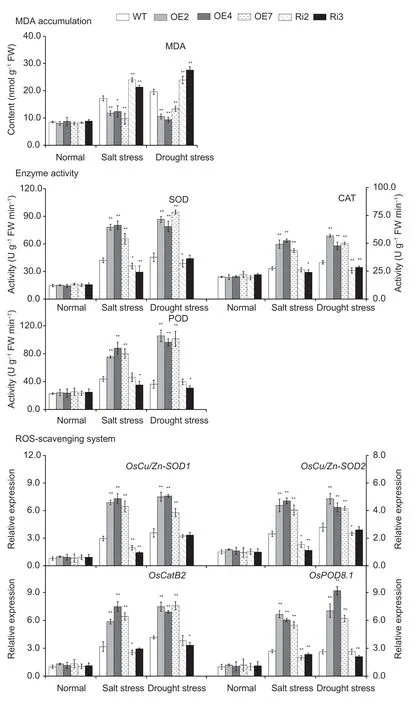
Fig.7 The content of malondialdehyde (MDA),the activities of superoxide dismutase (SOD),catalase (CAT),and peroxidase (POD) activities,and the expression of OsCu/Zn-SOD1,OsCu/Zn-SOD2,OsCatB2,and OsPOD8.1 genes in the leaves of OsMas1-OE plants,OsMas1-RNAi plants,and WT plants under salt and drought stresses.ROS,reactive oxygen species.Data are presented as mean±SE (n=3).* and ** indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
3.10.Overexpression of OsMas1 alters ion content in rice
The contents of K+and Na+in the leaves ofOsMas1-OE,OsMas1-RNAi,and WT plants under salt stress were measured.The results showed that K+content and K+/Na+ratio were significantly increased,while Na+content was significantly decreased inOsMas1-OE plants compared with WT andOsMas1-RNAi plants under salt stress (Fig.8).Next,we performed qRT-PCR assays to further analyze the expression levels of ion transportrelated genes after salt stress.The expression levels ofOsNHX1andOsNHX2,encoding Na+/H+exchangers,were increased inOsMas1-OE plants compared with WT andOsMas1-RNAi plants under salt stress (Fig.8).Our data showed that overexpression ofOsMas1leads to an increase of K+/Na+ratio under salt stress,indicating measurable improvement of salt tolerance in rice.
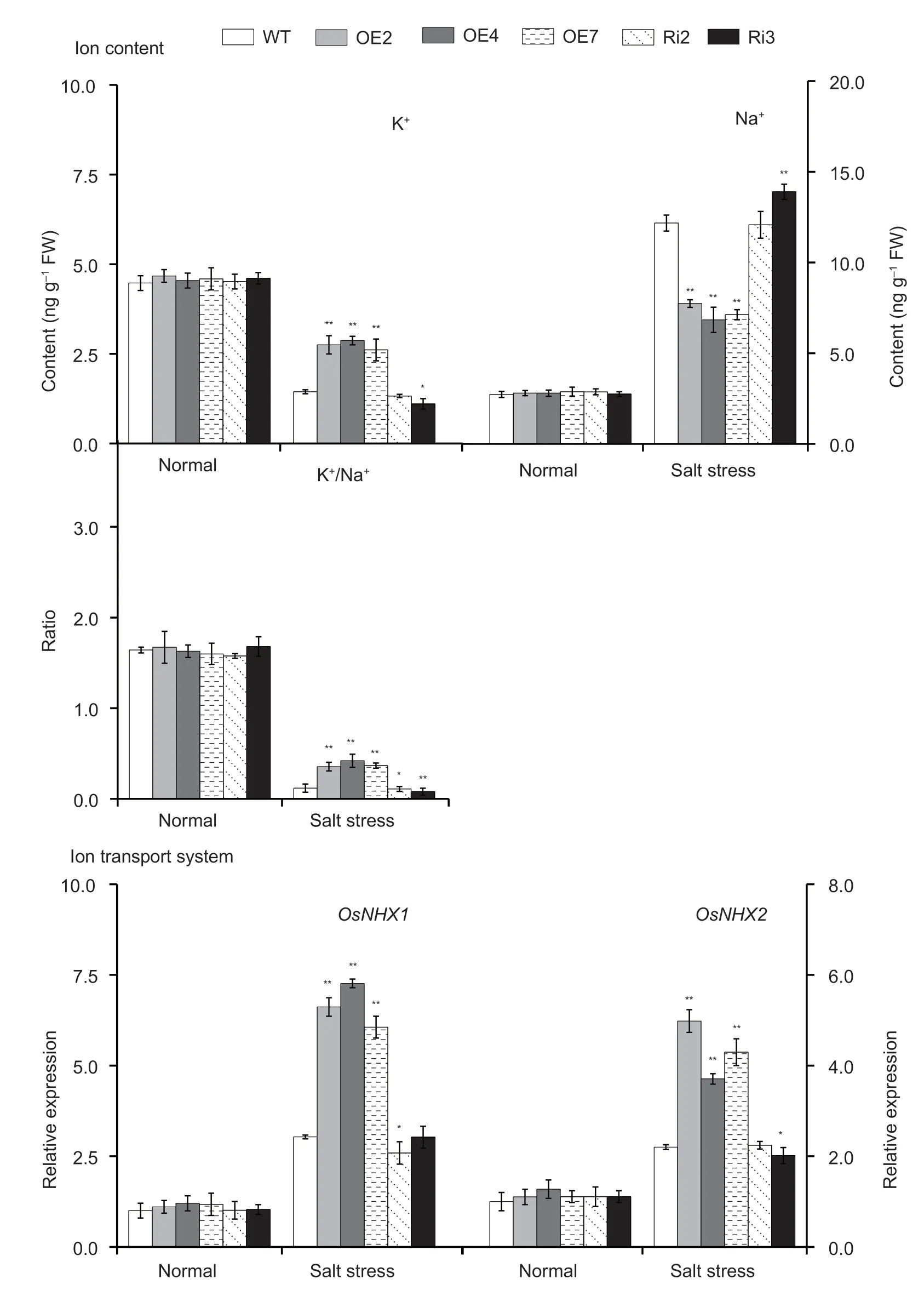
Fig.8 The content of K+ and Na+,the ratio of K+/Na+,and the expression of OsNHX1 and OsNHX2 genes in the leaves of OsMas1-OE,OsMas1-RNAi,and wild type (WT) plants under salt and drought stresses.Data are presented as mean±SE (n=3).* and **indicate a significant difference from that of WT at P<0.05 and P<0.01,respectively,by Student’s t-test.
4.Discussion
The maspardin protein was first identified as an intracellular binding protein for the cell surface glycoprotein CD4 and proposed to modulate CD4 stimulatory activity (Zeitlmannet al.2001).A nucleotide insertion (601insA) mutation in the maspardin gene resulted in the complicated form of hereditary spastic paraplegia known as Mast syndrome (Simpsonet al.2003).There was only one report on identifying theIbMasgene from sweetpotato at functional levels.The study of Liuet al.(2014) revealed that theIbMasgene was upregulated when exposed to salt stress and ABA treatment in a salt-tolerant sweetpotato line ND98,and theIbMasoverexpressing sweetpotato plants exhibited enhanced salt tolerance.However,little work is known about the regulatory functions of theOsMas1gene in regulating the abiotic stress tolerance of rice.
This study isolated a maspardin gene from the rice cultivar Yangjing 805.Sequence analysis showed that the protein contains a typical maspardin domain and thus is named OsMas1 (Fig.1-A).BLAST analysis indicated that OsMas1 protein exhibited 58.41 and 62.59%amino acid identity to an α/β-hydrolase superfamily in sweetpotato (AII99394) andArabidopsis(NP_192960).There was no report aboutArabidopsismutant for NP_192960.Here,we revealed the biological function and molecular regulatory mechanism ofOsMas1.The expression of theOsMas1gene was up-regulated by mannitol,PEG6000,and NaCl stresses and ABA treatment (Appendix B).Therefore,it is thought thatOsMas1may play an important role in enhancing the resistance of plants to abiotic and biotic stresses by the ABA-responsive signaling pathway.We further produced the transgenic plants of the ricecv.Zhonghua 11 by overexpressing and interfering with theOsMas1gene,respectively.We then found thatOsMas1-OE plants exhibited enhanced salt and drought tolerance.In contrast,OsMas1-RNAi plants showed decreased tolerance to salt and drought stresses compared with WT plants (Fig.2).It is thought that theOsMas1gene plays a vital role in response to salt and drought stresses of rice and can be directly used in the genetic engineering of crops for tolerance to abiotic stresses.
4.1.Os Mas 1 promotes salt and drought tolerance by activating ABA biosynthesis and signaling
As a central regulator of abiotic stress responses,endogenous ABA can participate in the regulation of a variety of physiological responses of plants to abiotic stresses,such as drought,salt,and chilling stresses,and regulate the expression of ABAdependent abiotic stress tolerance-responsive genes(Zhu 2002;Sripinyowanichet al.2013;Zhaiet al.2016;Liet al.2017,Li Get al.2020a).ABA-dependent stress-responsive genes,suchasOsRAB16C,OsRAB16D,OsRAB21,andOsLEA3,are well known for their involvement in response to salt and drought stresses of rice (Zhanget al.2014;Jianget al.2016;Liet al.2017,2018,2019).The expression level ofIb Maswas induced by ABA treatment,speculating that this gene may regulate sweetpotato salt stress response in an ABA-dependent manner(Liuet al.2014).In the present study,OsMas1-OE plants exhibited enhanced sensitivities,whileOsMas1-RNAi plants showed less sensitivity to exogenous ABA treatment,indicating thatOsMas1might be a positive regulator of ABA signaling in rice (Fig.3).The enhanced activity of NCED and up-regulated expression ofOsNCED4andOsNCED5and significant increase of endogenous ABA content was observed inOsMas1-OE plants compared with WT plants andOsMas1-RNAi plants under salt and drought stresses(Fig.4).Further observation showed that transcript levels of ABA-responsive signaling pathway genesOsRAB16C,OsRAB16D,OsRAB21,andOsLEA3were markedly upregulated inOsMas1-OE plants (Fig.4).The differences in salt and drought tolerance betweenOsMas1-OE,OsMas1-RNAi and WT plants might be,at least in part,due to the reinforced expression ofOsRAB16C,OsRAB16D,OsRAB21,andOsLEA3and possibly other contributing stress-responsive genes inOsMas1-OE plants.The results suggest that the enhanced salt and drought tolerance exhibited byOsMas1overexpression might be conferred by the coordinated work of these downstream functional genes.It is suggested that overexpression ofOsMas1enhances the tolerance to salt and drought stresses due to the up-regulation of genes involved in ABA biosynthesis,which increases the production of ABA as a signaling molecule and further the expression of ABA-responsive genes.
4.2.OsMas1 enhances salt and drought tolerance by regulating osmotic adjustments
Proline accumulation can enhance salt and drought tolerance in many plant species (Krasensky and Jonak 2012;Zhanget al.2019).ABA,salt,and drought treatments induced the expression ofOsP5CS,and the conferred stress tolerance resulted from an up-regulated expression ofOsP5CS,which increased proline content in rice plants (Xianget al.2007;Sripinyowanichet al.2013;Jianget al.2016;Liet al.2018).P5CS,catalyzing proline biosynthesis,is critical for enhanced salt and drought tolerance (Jianget al.2016).The up-regulation ofP5CShas been shown to increase proline content,resulting in enhanced salt and drought tolerance (Krasensky and Jonak 2012;Liuet al.2014;Jianget al.2016;Zhaiet al.2016;Kanget al.2018).As shown in Fig.5,OsMas1-OE plants exhibited the up-regulated expression levels ofOsP5CS1andOsP5CS2,increased activity of P5CS,and enhanced proline content under salt and drought stresses.More proline accumulation inOsMas1-OE plants might maintain the osmotic balance between the intracellular and extracellular environments and protect membrane integrity,resulting in enhanced salt and drought tolerance(Liuet al.2014;Zhaiet al.2016;Wanget al.2016;Zhanget al.2019;Li Get al.2020;Donget al.2022).Thus,it is suggested that accumulated ABA levels might up-regulate the expression of proline biosynthesis geneOsP5CS,which increases proline production,leading to enhanced salt and drought tolerance in rice.
Salinity and drought stresses perturb plant water uptake in leaves and disrupt the osmotic,ionic,and nutrient balances,leading to quick response in stomatal conductance.These stresses affect photosynthetic electron transport and the activities of enzymes for carbon fixation (Parida and Das 2005;Tuteja 2007;Liuet al.2014).The damaging effects of singlet oxygen and hydroxyl radicals on PSII can be reduced by proline in isolated thylakoid membranes (Aliaet al.1997;Liuet al.2014;Zhaiet al.2016;Zhanget al.2019).Proline protects PSII photofunctions against photodamage,which gets accelerated in plants under salt and drought stresses(De Rondeet al.2004;Liuet al.2014;Zhaiet al.2016;Huet al.2022;Zhaoet al.2022).In our study,OsMas1-OE plants exhibited enhanced photosynthesis capacity compared with WT andOsMas1-RNAi plants under salt and drought stresses (Fig.6).Also,the expression levels ofOsrbcL,OspsaA,OspsbA,andOsPRKgenes related to photosynthesis system were up-regulated inOsMas1-OE plants (Fig.6).The less affected photosynthesis ofOsMas1-OE plants could be explained by that the accumulated more proline inOsMas1-OE plants provides protection against photoinhibition under salt and drought stresses (Zhaiet al.2016;Kanget al.2018),similar to the results reported by Liuet al.(2014),in which theIbMas-overexpression sweetpotato plants exhibited enhanced photosynthesis performance.It is thought that overexpression ofOsMas1enhances photosynthesis capacity due to the up-regulation of photosynthesis system-related genes,leading to enhanced salt and drought tolerance.
The plantNHXgene family encodes Na+/H+antiporters,which are crucial for salt tolerance,potassium homeostasis,and cellular pH regulation (Qiu 2012;Huanget al.2018).A higher K+/Na+ratio can minimize Na+toxicity under salt stress,and it is generally accepted that maintenance of K+/Na+homeostasis is an important aspect of salt tolerance (Yueet al.2012).InArabidopsis,it has been proved that expression ofAtNHX1andAtNHX2increased in response to high salt stress through an ABA-dependent signaling pathway (Yokoiet al.2002).OsNHX1andOsNHX2transcripts accumulated in response to ABA in rice (Fukudaet al.2011).Overexpression ofOsNHX1andOsNHX2enhances salt tolerance in rice plants (Fukudaet al.2004;Liuet al.2010;Tenget al.2017).In the present study,Na+content was lower,and K+content and K+/Na+ratio were significantly higher inOsMas1-OE plants than in WT andOsMas1-RNAi plants under salt stress (Fig.8).qRT-PCR assays revealed that the expression levels ofOsNHX1andOsNHX2were up-regulated inOsMas1-OE plants compared with WT andOsMas1-RNAi plants (Fig.8).Thus,it is thought that high levels of ABA might upregulate the expression levels ofOsNHX1andOsNHX2,which increased K+/Na+ratio,indicating measurable improvement of salt tolerance.
4.3.OsMas1 improves salt and drought tolerance by adjusting ROS scavenging and balance
Under abiotic stresses,increased ROS production leads to oxidative stress in plants (Hussainet al.2019;Zhuet al.2020;Miaoet al.2021;Huet al.2022).Higher ROS levels induce lipid peroxidation in plants and cause injury to cell membranes (Zhaoet al.2018;Hussainet al.2019).MDA,a reflection of cellular membrane degradation or dysfunction,is an important intermediary agent in ROSscavenging (Zhuet al.2020).It is important to maintain a stronger ROS-scavenging ability under salt and drought stresses to alleviate the induced oxidative damage,especially in plant leaves where photosynthesis is dramatically impacted (Gill and Tuteja 2010).The higher ability of ROS-scavenging enzymes decreased overaccumulated ROS levels,leading to enhanced salt and drought tolerance (Apel and Hirt 2004;Jianget al.2016).SOD,CAT,and POD play an important role in scavenging ROS by detoxifying H2O2andinto water and stable oxygen,leading to enhanced stress tolerance (Wanget al.2018;Zhuet al.2020).In this study,OsMas1-OE plants exhibited lower levels of DAB and NBT staining,H2O2,and MDA contents compared with WT plants andOsMas1-RNAi plants under salt and drought stresses(Appendices H and I;Fig.7).This indicates a putative role ofOsMas1in ROS-scavenging and enhancing salt and drought tolerance.We also found that the systematic up-regulation of ROS scavenging genes (OsCu/Zn-SOD1,OsCu/Zn-SOD2,OsCatB2,andOsPOD8.1) and significantly increased antioxidant enzyme (SOD,CAT,and POD) activities were observed inOsMas1-OE plants under salt and drought stresses (Fig.7).Therefore,the improved salt and drought tolerance of the transgenic rice plants might be due,at least in part,to the enhanced ROS scavenging capacity (Liuet al.2014;Zhaiet al.2016;Wanget al.2018;Li Get al.2020).It has been reported that proline is an effective scavenger of singlet oxygen and hydroxyl radicals (Liuet al.2014;Zhaiet al.2016;Zhanget al.2019;Zhaoet al.2022).Our results support that more proline accumulation activates the ROS scavenging system,leading to enhanced salt and osmotic tolerance inOsMas1-OE plants (Zhaiet al.2016;Wanget al.2018;Li Get al.2020).
5.Conclusion
This study presented a report on the function ofOsMas1in regulating salt and drought tolerance in rice.OsMas1plays an essential role in regulating salt and drought tolerance by up-regulation expression of abiotic stressresponsive genes involved in ABA signaling,proline biosynthesis,ROS scavenging,photosynthesis,and ion transport pathways.Therefore,we suggest thatOsMas1may be helpful for enhancing plant tolerance to abiotic stresses.
Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province,China (BK20191483),the Natural Science Fund for Colleges and Universities in Jiangsu Province,China (20KJA180004),the Postgraduate Practice Innovation Program of Jiangsu Province,China (SJCX20_1339),the College Student Practice Innovation Program of Jiangsu Province,China(202111049104H,202211049133H and 202211049138H)and the Talent Introduction Research Project of Huaiyin Institute of Technology,China (Z301B16534).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- A 314-bp SlNE insertion in the ZNF2 promoter region may act as a repressor related to regulation of fat deposition in pigs
- An optimized protocol using Steedman’s wax for high-sensitivity RNA in situ hybridization in shoot apical meristems and flower buds of cucumber
- Characterization of subunits encoded by SnRK1 and dissection of combinations among these subunits in sorghum (Sorghum bicolor L.)
- The role of time preferences in contract breach: Evidence from Chinese poultry farmers participating in contract farming
- Optimal design of culling compensation policy under the African swine fever — Based on simulations of typical pig farms in China
- Drip fertigation and plant hedgerows significantly reduce nitrogen and phosphorus losses and maintain high fruit yields in intensive orchards
