The dynamic metabolic profile of Qi-Yu-San-Long decoction in rat urine using UPLC-QTOF-MSEcoupled with a post-targeted screening strategy
2022-12-02TingZhengYueZhaoRuijuanLiMengwenHuangAnZhouZegengLiHuanWu
Ting Zheng,Yue Zhao,Ruijuan Li,Mengwen Huang,An Zhou,Zegeng Li,Huan Wu
Key Laboratory of Xin'an Medicine,Ministry of Education,Anhui University of Chinese Medicine,Hefei,230038,China
ABSTRACT
Qi-Yu-San-Long decoction(QYSLD)is a traditional Chinese medicine that has been clinically used in the treatment of non-small-cell lung cancer(NSCLC)for more than 20 years.However,to date,metabolicrelated studies on QYSLD have not been performed.In this study,a post-targeted screening strategy based on ultra-performance liquid chromatography coupled with quadrupole time-of-flight full information tandem mass spectrometry(UPLC-QTOF-MSE)was developed to identify QYSLD-related xenobiotics in rat urine.The chemical compound database of QYSLD constituents was established from previous research,and metabolites related to these compounds were predicted in combination with their possible metabolic pathways.The metabolites were identified by extracted ion chromatograms using predicted m/z values as well as retention time,excimer ions,and fragmentation behavior.Overall,85 QYSLD-related xenobiotics(20 prototype compounds and 65 metabolites)were characterized from rat urine.The main metabolic reactions and elimination features of QYSLD included oxidation,reduction,decarboxylation,hydrolysis,demethylation,glucuronidation,sulfation,methylation,deglycosylation,acetylation,and associated combination reactions.Of the identified molecules,14 prototype compounds and 58 metabolites were slowly eliminated,thus accumulating in vivo over an extended period,while five prototypes and two metabolites were present in vivo for a short duration.Furthermore,one prototype and five metabolites underwent the process of“appearing-disappearing-reappearing”in vivo.Overall,the metabolic profile and characteristics of QYSLD in rat urine were determined,which is useful in elucidating the active components of the decoction in vivo,thus providing the basis for studying its mechanism of action.
Keywords:
Qi-Yu-San-Long decoction
Post-targeted screening strategy
Dynamic metabolic profile
UPLC-QTOF-MSE
1.Introduction
Research on drug metabolism is an important avenue to understand the fate of Chinese herbal medicine and traditional Chinese medicine(TCM)prescriptions in vivo,thus being valuable in illuminating the pharmacological effects of these medicines[1-3].However,because of the complexity of the chemical components of TCM and the biotransformation of these components in vivo over time,investigations into their associated metabolic pathways have always been an extremely challenging task[4,5].To overcome this issue,it is necessary to establish a highly versatile,high-resolution,and highly selective assay technique to analyze biological samples at multiple time intervals following drug administration.The analytical method should have the capability to determine when compounds are transformed,which metabolic pathways are involved at a specific time,and any associated potential activity and/or toxicity from the investigated compounds and the corresponding metabolites.
Qi-Yu-San-Long decoction(QYSLD)is a TCM prescription that has been clinically used for the effective treatment of non-smallcell lung cancer(NSCLC)for more than 20 years[6].QYSLD is composed of Astragali Radix(Huangqi),Solanum nigrum L.(Longkui),Scolopendra(Tianlong),Pheretima(Dilong),Euphorbia helioscopia L.(Zeqi),Hedyotis diffusa Willd.(Baihuasheshecao),Curcumae Rhizoma(Ezhu),Coicis semen(Yiyiren),Polygonati Odotati Rhizoma(Yuzhu),and Fritillaria cirrhosae Bulbus(Chuanbeimu)in various proportions.Previous pharmacological experiments have shown that the inhibitory effects of QYSLD on NSCLC are related to its regulation,at a molecular expression level,of the Wnt/β-catenin, P13K/Akt/mTOR, and TLR4/MyD88/NF-κB pathways[7-9].A previous metabolomics study demonstrated that QYSLD exerts an inhibitory effect against NSCLC both in vivo and in vitro,and the potential biomarkers associated with the therapeutic effect were identified from the perspective of endogenous metabolites[10].The inhibitory effect of QYSLD on NSCLC in vivo is closely related to its chemical composition and metabolites;therefore,it is important to understand the dynamic metabolic changes in the chemical composition of the decoction in vivo.Urine is considered as an ideal biological sample for studying metabolic patterns in vivo because it is easy to collect,does not puncture the skin like in the case of blood sample collection,and generally contains a relatively high concentration of drugs and their metabolites following renal elimination.In addition,chemical components of TCM can be determined from urine samples following renal excretion,thus reflecting the residence of the compounds and their metabolites in the body,which are crucial parameters for assessing the safety of these medicines.
The rapid development of high-resolution mass spectrometry(HRMS)coupled to various analyzers,including orbitrap mass spectrometry(MS),quadrupole-time-of-flight(QTOF)MS,linearion-trap MS,and ion-trap-TOF MS,has led to its broad application in drug impurity analyses and metabolic studies because of its enhanced capacity of measuring the exact mass of analytes and fragmentations[11-14].Among these HRMS methods,Q-TOF MS combined with ultra performance liquid chromatography(UPLCQTOF-MS)has excellent separation performance,high resolution,high sensitivity,a wide dynamic scan range,and can be used for the rapid identification and semiquantitative analysis of complex components[15-19].It has become the most commonly used technique for the analysis of various components of herbal medicine[20]and TCM prescriptions[21-23].Nevertheless,metabolic profiling of TCM prescriptions in vivo remains extremely challenging,mainly because of the presence of a large number of chemical components with diverse structural features[24],complex transformation reactions in vivo[25],and interference of endogenous substances[26].Owing to this,establishing an analytical strategy that relies on highselectivity and high-resolution analytical instruments is crucial for overcoming these challenges.
Comprehensive and efficient characterization of biological samples after the administration of TCM requires highly accurate and adequate precursor ion scanning,as well as corresponding secondary fragment ion information to facilitate structural analysis[11].Typically,MS-based analytical methods are designed to capture targets of interest during data acquisition and post-acquisition data interpretation.MS data acquisition is divided into dataindependent acquisition(DIA)and data-dependent acquisition(DDA)[27].DDA methods based on HRMS include the mass defect filter(MDF),product ion filter(PIF),and neutral loss filter(NLF)[28].PIF detects metabolites based on predicted product ions,whereas NLF detects metabolites based on predicted neutral loss fragments.Owing to the high selectivity of PIF and NLF,these data mining tools have been applied to identify trace metabolites in vivo[29].However,there are limitations to the detection of metabolites that do not produce significantly predictable product ions or neutral loss fragments[30].Compared with PIF and NLF,MDF relies on the deviation between the m/z value of an ion and its nearest whole number(accurate to four decimal places).Some welldefined narrow windows of the MDF template effectively remove most of the interference and allow researchers to obtain the target metabolite accurately[11].However,unknown metabolites that contain uncommon groups are beyond the range of predicted quality defects,making MDF incapable of detecting these target compounds.In addition,MDF does not consider the characteristic fragmentation patterns of interference,which can lead to falsepositive results[31].
In contrast to DDA,DIA does not require pre-selection of compounds in the sample but directly collects the MS information of all compounds separated by the chromatographic column.MSE,one of the DIA techniques,is a data acquisition method that can be used to alternately scan,by “low collision energy”and“high collision energy”,and to obtain highly accurate information of parent ions and fragment ions,as well as correlate and assign them based on their chromatographic behavior[32].Based on the full spectral information that can be collected using UPLCQTOF-MSE,we developed a post-targeted screening strategy in this study.We applied the strategy to predict the metabolites from QYSLD based on the biotransformation pathways of the different constituents of the prescription and to obtain the molecular formula and exact mass of the predicted metabolites.Following this,ion flow was determined using the predicted m/z values in conjunction with characteristic fragment ions and compound retention time(tR)to identify the predicted metabolites.Subsequently,we successfully characterized the prototype components and metabolites of QYSLD and delineated their changes in urine samples from rats,thus providing valuable information for further studies on the potentially active or toxic constituents of QYSLD.
2.Experimental
2.1.Materials and reagents
QYSLD was prepared from the procedure outlined in a previous study[10].The specific procedural steps are outlined in the Supplementary data.Each medicinal product that was used for the preparation of QYSLD was identified by Professor Qingshan Yang of Anhui University of Chinese Medicine,Hefei,China.
Calycosin (MUST-19120901,99.8%,1.42 g/cm3,molecular weight:284.0685)and E-p-coumaric acid(MUST-20050603,99.9%,1.14 g/cm3,molecular weight:164.0473)of reference standards were purchased from Manster Biotechnology Co.,Ltd.(Chengdu,China).Peimisine(B20082,98%,1.17 g/cm3,molecular weight:427.3086)was purchased from Yuanye Biotechnology Co.,Ltd.(Shanghai,China),and Milli-Q ultra-pure water(Millipore,Billerica,MA,USA)was used for all experimental procedures.Acetonitrile and methanol(LC-MS grade)were obtained from Merck(Darmstadt,Germany),and formic acid(Chromatographic grade)was obtained from Sigma-Aldrich(St.Louis,MO,USA).A KQ-500DB CNC ultrasonic cleaner and RE-3000A rotary evaporator were obtained from Ultrasonic Instrument Co.,Ltd.(Kunshan,China)and Shanghai Yarong Biochemical Instrument Factory (Shanghai,China),respectively.
2.2.Animal experiments
Twelve male Sprague-Dawley(SD)rats(200-250 g)of specific pathogen-free(SPF)grade were fed adaptively for seven days with access to chow and drinking water.All rats were housed in metabolic cages for three days before administration of the test or control product.The 12 rats were then divided into two groups:the QYSLD group and the blank group.All rats were fasted for 12 h before intragastric administration.The rats in the QYSLD group were administered QYSLD suspension twice a day for three consecutive days at a dose of 25 mL/kg per day.Urine was collected between 0 and 2,2-4,4-8,8-12,and 12-24 h after intragastric administration of QYSLD.The rats in the blank group were administered the suspension vehicle(distilled water;25 mL/kg)twice daily for three days.Urine samples from animals in the blank group collected at each time point were mixed to serve as a blank control.After sample collection,the urine was centrifuged(3,000 r/min,10 min,4°C)and the supernatant was frozen at-80°C.All protocols were approved by the Animal Ethics Committee of Anhui University of Chinese Medicine,Hefei,China.The procedures involving animal research conformed to the animal ethics procedures and Guide for the Care and Use of Laboratory Animals[33].
2.3.Pretreatment of urine samples
Urine samples(200μL)were placed in 2.0 mL Eppendorf tubes.Next,600μL of cold methanol was added and samples were swirled for 15 s and then centrifuged(12,500 r/min,10 min,4°C).Subsequently,the supernatant was transferred and dried under N2,and the residue was redissolved in 200μL of methanol,vortexed,and centrifuged(12,500 r/min,10 min,4°C).The supernatant was then placed in an injection vial for analysis.
2.4.Instruments and analytical conditions
2.4.1.Chromatography
Urine samples were separated by chromatography using an ACQUITY I-Class UPLC system(Waters Corporation,Milford,MI,USA)on a Waters ACQUITY UPLC BEH C18analytical column(100 mm×2.1 mm,1.7μm).The chromatographic parameters were set as follows:column temperature,48°C;flow rate,0.2 mL/min;temperature of the autosampler,4°C;mobile phase,solvent A(0.1% formic acid in water)and solvent B(acetonitrile).Linear gradient was applied for elution as follows:0-7 min,3%-15% B;7-11 min,15% B;11-21 min,15%-25% B;21-26 min,25%-35% B;26-36 min,35%-55% B;36-45 min,55%-73% B;45-51 min,73%-85% B;51-56 min,85%-95% B;56-61 min,95% B;61-62 min,95%-3% B;and 62-65 min,3% B.
2.4.2.MS
A Waters Xevo G2-XS QTOF/MS detector(Waters Corporation,Milford,MI,USA)was applied for detection using an electrospray ion source.The mass spectrometric parameters were as follows:capillary voltage,2.5 kV(negative ion mode)and 3.0 kV(positive ion mode);sampling cone voltage,40.0 V;ion source temperature,120°C;desolvent gas flow rate,600 L/h;conical gas flow rate,40 L/h;desolvent temperature,350°C.The low collision energy of the MSEmode was 6 eV,the high energy was 20-35 eV,and the scanning range was m/z 50-1,200.Leucine enkephalin(200 pg/mL,10μL/min)was used as a real-time correction fluid.
2.5.Post-targeting analysis strategy
Due to the large number of chemical compounds with diverse structural features in TCM prescriptions,complex transformation reactions in vivo,and the interference of endogenous substances,we established a post-targeted screening strategy to identify QYSLD-related xenobiotics in rat urine samples(Fig.1).The analysis strategy was as follows.
First,a chemical compound database for QYSLD was established.Information on 166 constituents(name,formula,structure,retention time,excimer ion,and characteristic fragment ion)of QYSLD was collected,summarized based on our previous study[34],and entered into an Excel spreadsheet.
Second,prototype compounds were rapidly identified.After data collection using the UPLC-QTOF-MS with an MSEfunction,the ion flow was extracted at a MassLynx 4.1 workstation according to the QYSLD chemical compound database,and the associated retention time and excimer ion were obtained.The prototype compounds were then determined based on MS/MS ions in the database.
The third step involved the prediction and analysis of metabolites.After administration of TCM prescriptions,the structures of chemical compounds can be changed following the actions of a variety of drug-metabolizing enzymes,especially hepatic enzymes.The biotransformation of drugs in vivo includes phase I and phase II reactions.Phase I reactions are mainly the functional reactions of drugs under the catalysis of enzymes,which introduce or expose polar groups of these molecules(e.g.,oxidation reaction that can introduce hydroxyl groups into drug molecules,and hydrolysis reaction that fully exposes the carboxyl and hydroxyl groups in the fully exposed drug molecules).Phase II is usually called the binding reaction,in which the polar groups inherent in the compound or produced during the phase I metabolism stage are combined with endogenous components(e.g.,glucuronic acid,and glutathione)in vivo and then excreted from the body or reabsorbed back into the body.Based on the chemical constituents of QYSLD identified in vitro,combined with their possible metabolic pathways,we predicted the probable corresponding metabolites and summarized the molecular formula and exact mass of the projected molecules.For example,a precursor compound(CxHyOzNm)can undergo phase I metabolic reactions in vivo,such as hydrolysis reaction,to produce a metabolite(CxHy+2Oz+1Nm),and then undergo a phase II metabolic reaction,such as glucuronidation,to produce another metabolite(Cx+6Hy+10Oz+7Nm).Next,the ion flow is then extracted using the predicted m/z values in combination with characteristic fragment ions and the retention time,ultimately identifying the metabolites of this compound.
Last,false positive results were eliminated.The chromatogram and mass spectra data of the identified prototype compounds and metabolites in the urine samples of the QYSLD group were compared with those of the blank group.If samples from the blank and QYSLD groups gave the same predicted m/z at the same retention time,the metabolite derived from the corresponding predicted m/z was excluded as a false-positive result.
3.Results and discussion
3.1.Summary analysis of QYSLD metabolism
The total ion chromatograms(TICs)of rat urine samples obtained by UPLC-QTOF-MSEin the electrospray ionization positive(ESI+)and ESI-modes are shown in Fig.S1.According to the posttargeted screening strategy based on UPLC-QTOF-MSE,85 QYSLD-related xenobiotics were identified in rat urine,including 20 prototype compounds(P1-P20)and 65 metabolites(M1-M65).Specific information on the prototype compounds and metabolites of QYSLD in rat urine is shown in Tables S1 and S2,respectively.
3.2.Characterization of prototype compounds of QYSLD in rat urine
In this study,after intragastric administration of QYSLD to rats,urine was collected for 24 h to fully identify the prototype compounds of the prescription,with a total of 20 prototype compounds detected,including terpenoids(P1,P2,P3 P5,P7,P16,P18,and P19),flavonoids(P10,P13,and P17)and their glycosides(P8 and P14),alkaloids(P11 and P12),anthraquinones(P15 and P20),coumarins(P6 and P9),and quinoline(P4),of which three prototype compounds(E-p-coumaric,peimisine,and calycosin)were identified using reference standards,as shown in Figs.S2-S4.For example,a prototype compound with a retention time of 9.2 min that exhibited a molecular ion at m/z 163.0388[M-H]-was found to have the molecular formula C9H8O3.This molecular formula was consistent with that of E-p-coumaric acid in the self-built database.In addition,daughter ions at m/z 119.0492[M-H-COO]-,117.0328[M-H-COO-2H]-,and 93.0329[M-H-COO-C2H2]-were observed in the MS/MS spectrum(Fig.S2).Furthermore, the fragment ions of P6 were similar to those of E-p-coumaric acid.Consequently,P6 was identified as E-pcoumaric acid according to its excimer and fragment ions when compared to the self-built database.The other 19 prototype components were identified in a similar manner.

Fig.1.Post-targeted screening strategy for the characterization of Qi-Yu-San-Long decoction(QYSLD)-related xenobiotics in rat urine samples.The red portions represent the molecular formula.
The ion chromatograms of the 20 prototype compounds were extracted(Fig.2).P1,P3,P4,P6,P7,P9,P13,P16,P17,P18,and P19 were detected during the entire 24 h investigation period.These compounds were slowly eliminated in vivo and thus accumulated during this period.Among the prototype compounds,P2 and P15 were detected at 2 h;P14 at 4 h but was not detected at 24 h;P11 and P20 were detected at 24 h and were not present at 12 h;P5 and P12 were present in the first 8 h;P8 was detected at 12 h but was not detected at 24 h;and P10 was detected between 2 and 4 h and 12-24 h.It is worth reporting that P10 underwent“appearancedisappearance-appearance”in vivo.This phenomenon may be related to the enterohepatic circulation of the compound.
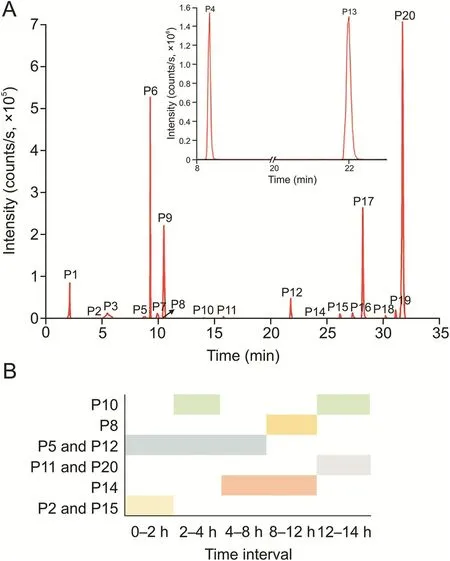
Fig.2.(A)Extracted ion chromatograms of prototype components of Qi-Yu-San-Long decoction(QYSLD)in rat urine and(B)their occurrence at different time periods.
3.3.Characterization of QYSLD metabolites in rat urine
Herein,urine samples were collected at different time periods(0-2 h,2-4 h,4-8 h,8-12 h,and 12-24 h)after the administration of QYSLD and were analyzed using UPLC-QTOF-MSE.According to the developed post-targeted screening strategy,65 metabolites of QYSLD were identified in rat urine,and were divided into various categories including terpenoid-related,flavonoidrelated,and coumarin-related.The extracted ion chromatograms of the 65 metabolites are shown in Fig.3.
3.3.1.Terpenoid-related metabolites
The terpene-related metabolites of QYSLD included iridoidrelated metabolites(M2,M10,M14,M24,M34,M38,M48,M49,M51,M56,M57,M58,and M63),sesquiterpene-related metabolites(M22,M31,M33,M37,M40,M44,and M45),and triterpene-related metabolites(M64 and M65).For iridoid-related metabolites,M2,M38,M51,M57,and M63 were deglycosylated metabolites derived from P1,P3,(E)-6-O-p-feruloyl scandoside methyl ester,(E)-6-O-pcoumaroyl scandoside methyl ester,and P7 constituents of QYSLD,respectively.In addition,M38 was found to undergo additional methylation in vivo to form M49.After the deglycosylation of iridoid glycosides,the polarity of the generated iridoid-related metabolites decreased,and their retention times in the C18column were prolonged.For example,M2 appeared at tR=2.5 min with an m/z of 227.0553[M-H]-,which was a 162 Da difference from the mass-charge ratio of deacetyl asperulosidic acid,and the retention time of deacetyl asperulosidic acid(tR=2.1 min)was shorter than that of M2.Combined with the post-targeting analysis strategy,we speculated that M2 was the deglycosylation product of deacetyl asperulosidic acid,according to the predicted m/z value.Moreover,under the high-energy collision of high-resolution mass spectrometry,major fragmentation ions were observed at m/z values of 209.0443[M-H-H2O]-,181.0337[M-H-H2O-CO]-,167.0423[M-H-CO2-O]-,165.0568[M-H-H2O-CO2]-,and123.0429[M-H-H2O-C3H2O3]-in the negative mode(Fig.4).These fragment ions are consistent with the characteristic fragments of deacetyl asperulosidic acid.Therefore,M2 was considered to be the deglycosylated metabolite of deacetyl asperulosidic acid.Other iridoid-related metabolites were characterized in the same way as described above;detailed information regarding this is shown in Table S2.
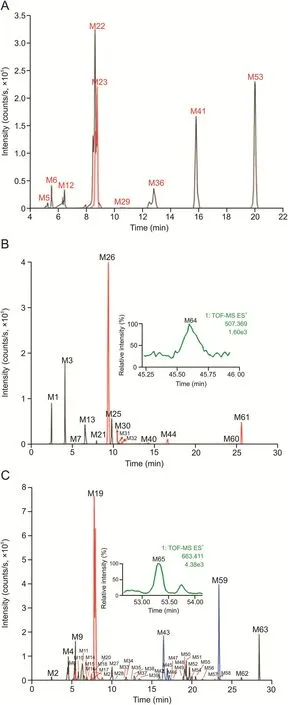
Fig.3.Extracted ion chromatograms of Qi-Yu-San-Long decoction(QYSLD)-related metabolites in rat urine.(A)Metabolites that could be detected in both electrospray ionization negative(ESI-)and positive ESI+modes;(B)metabolites that could be detected only in ESI+mode;and(C)metabolites that could be detected only in ESI-mode.
In the case of sesquiterpenes,loliolide,13-hydroxygermacrone,and bisacumol combined with glucuronic acid to form more polar compounds such as M22,M33,and M37.In addition,loliolide underwent acetylation reactions to generate M31 in vivo.As an example,M22 had a precursor ion[M-H]-at m/z 371.1334,and the main characteristic product ions of M22 were m/z 195.0643[M-H-C6H8O6]-,m/z151.0753[M-H-C6H8O6-CO2]-,m/z177.1262[M-H-C6H8O6-H2O]-,and m/z 179.0349[M-H-C6H8O6-O]-in ESI-mode(Fig.5).The precursor ion of M22 was 176 Da more than that of loliolide, and the characteristic fragments of M22 were similar to those of loliolide.Consequently,M22 was considered as a glucuronic acid conjugate of loliolide based on its related mass spectrometry cleavage behavior. Other sesquiterpene-related metabolites were characterized in the similar manner and detailed information regarding this is provided in Table S2.
In a previous report on the chemical constituents of QYSLD[34],astragalosides were the main triterpenoid glycosides.When we characterized the prototype components in the samples of rat urine,no triterpenoid saponins were detected.Saponins are generally considered to have low oral bioavailability[35,36].In addition,only two metabolites related to astragalosides(M64 and M65)were detected in this study,which showed a relatively poor recovery.During UPLC-QTOF-MSEanalysis,M64 had a precursor ion[M+H]+at m/z 507.3686,and its precise mass was 16 Da greater than that of aglycone astragaloside IV.Under high-energy bombardment,some fragment molecules were lost,producing m/z 487.2836[M+H-H2O-2H]+and 475.3345[M+H-2O]+fragments.Subsequently,M64 was considered to be the oxidation product of the aglycones of astragaloside IV.M65 had a precursor ion[M-H]-at m/z 663.4174,which was 162 Da less than that of astragaloside II,and produced characteristic fragments of m/z 647.4553[M-H-O]-and 621.4333[M-H-C2H2O]-.These two fragment ions were also characteristic fragments of astragaloside II.Therefore,M65 was considered as a deglycosylated product of astragaloside II.
3.3.2.Flavonoid-related metabolites
A large number of flavonoids and their associated glycosides are present in QYSLD[34].Consequently,their related metabolites were postulated to be the main components of the characterized urine samples in this study.Herein,16 flavonoid-related metabolites were identified,three of which(M28,M29,and M35)were related to rutin,four(M17,M39,M42,and M62)to kaempferol,six(M36,M41,M43,M46,M47,and M54)to calycosin,and three(M1,M23,and M53)to formononetin.A previous study suggested that the glycosidic bonds of flavonoid glycosides were easily broken and then cleaved off by Retro-Diels-Alder(RDA)to form fragment ions[37].As depicted in Fig.6,M29 was cleaved by RDA to form the characteristic fragment ions of m/z 121.0648[M-H-C8H4O5]-and 107.0480[M-H-C9H6O5]-.In addition to the RDA reaction,M29 also produced characteristic fragments of m/z 193.0348[M-H-C6H4O2]-and 165.0542[M-H-C6H4O2-CO]-.These ions are identical to the characteristic fragments of quercetin;therefore,M29 was identified as quercetin.Studies have shown that the main metabolic pathways of isoflavones are sulfation and glucuronidation[38,39],which is consistent with our obtained results.For example,M53 with m/z 443.0974[M-H]-appeared at 20.0 min and lost glucuronic acid(176 Da)to produce a fragment of m/z 267.0659[M-H-C6H8O6]-,which further underwent RDA to yield ions at m/z 135.0053[M-H-C6H8O6-C9H8O]-and 132.0191[M-H-C6H8O6-C7H3O3]-.M53 also produced other fragment ions at m/z 253.0493 [M-H-C6H8O6-CH2]-and 225.0555[M-H-C6H8O6-CH2-CO]-(Fig.S5).The ion at m/z 267.0659 and other fragment ions were the same as the excimer ion and the fragment ions of formononetin in the self-built library;therefore,M53 was identified as a glucuronic acid conjugate of formononetin.M43 appeared at 16.5 min,and the neutral loss of SO3(80 Da)produced a fragment ion with a m/z of 283.0588[M-H-SO3]-.In the MS/MS spectrum,ions at m/z values of269.0391[M-H-SO3-CH2]-,241.0439[M-H-SO3-CH2-CO]-,239.0340[M-H-SO3-CO-O]-, 148.0150 [M-H-SO3-C7H3O3], and 135.0433[M-H-SO3-C9H8O2]were observed(Fig.S6).The ion at m/z 283.0588 and other fragment ions were the same as the excimer ion and fragment ions of calycosin in the self-built library.Owing to this,M43 was considered to be a sulfated compound of calycosin.In addition,flavonoids were acetylated under the catalysis of acyltransferase to produce fewer polar metabolites,such as M35,M42,M46,and M54.

Fig.4.Product ions obtained by ultra-performance liquid chromatography coupled with quadrupole time-of-flight full information tandem mass spectrometry(UPLC-QTOF-MSE)in electrospray ionization negative(ESI-)mode and fragmentation pathways of M2.
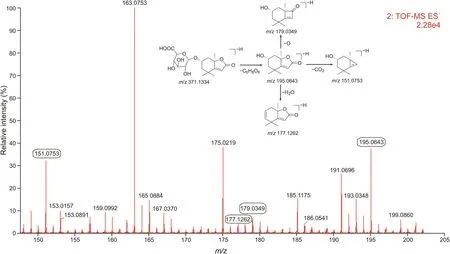
Fig.5.Product ions obtained by ultra-performance liquid chromatography coupled with quadrupole time-of-flight full information tandem mass spectrometry(UPLC-QTOF-MSE)in electrospray ionization negative(ESI-)mode and fragmentation pathways of M22.
3.3.3.Coumarin-related metabolites
Based on post-targeting analysis,17 coumarin-related metabolites were identified in rat urine,which were considered to be related to ferulic acid(M8,M9,M16,M19,and M27),E-pcoumaric acid(M4,M11,M13,M25,M26,and M32),and scopoletin(M5,M6,M7,M12,M15,and M18).These metabolites maintained the related structures of their precursors,and their cleavage pathways were essentially the same.M25(tR=9.8 min)was detected at m/z 167.0720[M+H]+in ESI+mode,and under further high-energy bombardment,generated ions of m/z 123.0832[M+H-COO]+,121.0667[M+H-COO-2H]+,107.0510[M+H-COO-O]+,and 105.0702[M+H-COO-O-2H]+(Fig.S7).These fragment ions were consistent with those of E-p-coumaric acid in the self-built library;therefore,M25 was regarded as the reduction product of E-p-coumaric acid according to the observed mass spectrometry cleavage behavior.M5 was detected in both ESI+and ESI-modes,and its molecular formula(C10H10O5)was different from that of scopoletin by an H2O(18 Da)molecule.The excimer ion peak was at m/z 209.0443[M-H]-,and the fragment ions had m/z values of 165.0516[M-H-COO-]-,147.0283[M-H-COO-H2O]-,and117.0505[M-H-COO-H2O-OCH2]-(Fig.S8).The ions at m/z 147.0283[M-H-COO-H2O]-and117.0505[M-H-COO-H2O-OCH2]-were characteristic fragments of scopoletin.Therefore,M5 was identified as scopoletin hydrolysate.The other coumarin-related metabolites were characterized in the same manner and detailed information regarding this is provided in Table S2.
3.3.4.Other metabolites
In addition to the aforementioned metabolites,other QYSLD metabolites identified from rat urine samples included two alkaloidrelated metabolites(M30 and M60),five anthraquinone-related metabolites(M50,M52,M55,M59,and M61),one lignan-related metabolite(M20),and two quinoline-related metabolites(M3 and M21).These metabolites were identified by combining cleavage patterns and the exact quality of their respective precursor compounds.For example,M52(tR=19.7 min)showed[M-H]-at m/z 475.0900,and the characteristic fragmentation of M52 at m/z 299.0565 was caused by the loss of 176 Da(glucuronic acid),which was further broken into m/z 285.0798[M-H-C6H8O6-CH2]-and m/z 271.0769[M-H-C6H8O6-CO]-.These ions are consistent with the characteristic fragments of 2,6-dihydroxy-1-methoxyanthraquinone.Thus,M52 was characterized as a glucuronic acid conjugate of 2,6-dihydroxy-1-methoxyanthraquinone.M3 and M59 were identified in a similar manner.
3.4.Dynamic metabolic profile of QYSLD in rat urine
To further explore the detailed metabolic profile of QYSLD constituents,urine samples were collected at different time intervals and analyzed.As shown in Fig.7,M14 and M60 were present between 2-24 h and 8-24 h,respectively.This phenomenon indicated that the two metabolites were slowly absorbed and eliminated.Unlike the other metabolites,M64 and M65 were detectable for only 2 h.These results show that these two metabolites were rapidly eliminated in vivo.In addition,the following were observed:M6,M10,M12,M17,M18,M21,M26,M29,M32,M35,M37,M47,M48,M52,M53,and M56 were present between 0 and 8 h;M4,M24,M43,and M62 between 0 and 12 h;M7,M8 and,M15 between 0-2 h and 4-8 h;M30 between 0-2 h and 4-12 h;M9 between 0-8 h and 12-24 h.All the other metabolites were detectable during the entire 24 h period.
It is worth noting that among the 65 metabolites that were characterized,49 were related to the components present as prototypes of QYSLD in vivo(Fig.8).
After the overall biotransformation of the investigated products,calycosin(P13)was present as a reduction product(M47)between 0 and 8 h,a sulfate-conjugated compound(M43)between 0 and 12 h,and a glucuronic acid conjugate(M41)during the 24 h period.M36(demethylation and glucuronidation),M46(demethylation and acetylation),and M54(reduction and acetylation)were observed metabolites of calycosin produced after phase I and phase II reactions in vivo.These three metabolites were also present between0 and24h(Fig.9).The detection response of calycosin and its glucuronic acid conjugate(M41)in terms of mass spectrometry was higher than that of the other metabolites.These results suggested that calycosin was mainly eliminated as the prototype molecule and its glucuronic acid conjugate after 24 h.Similar to calycosin,the metabolite(M23)of formononetin after demethylation and glucuronidation was detected during the 24 h period.In contrast to calycosin,the glucuronic acid conjugate of formononetin(M53)was present between 0 and 8 h.This phenomenon indicated that the glucuronic acid conjugate of formononetin was more easily eliminated in vivo than that of calycosin.Another formononetin reduction-metabolite(M1)was present between 0 and 24 h(Fig.S9).Compared to the reduction product of calycosin,M1 was eliminated slowly and thus accumulated for a long time in vivo.
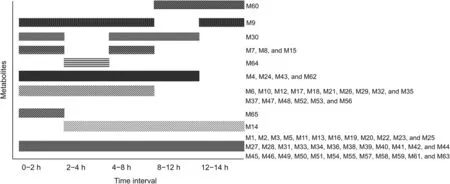
Fig.7.Dynamic changes of Qi-Yu-San-Long decoction(QYSLD)metabolites in rat urine.
Furthermore,E-p-coumaric acid(P6)and scopoletin(P9)were transformed into various metabolites in vivo.Decarboxylation and acetylation metabolites(M26 and M32)of E-p-coumaric acid were present between 0 and 8 h.The metabolite M11,which was formed by the further sulfation of M26,was present during the 24 h period.The sulfated and reduced products of E-p-coumaric acid(M13 and M25)were also present during this period(24 h).Another metabolite(M4),the glucuronic acid conjugate of E-p-coumaric acid,was present between 0 and 12 h(Fig.S10).In the first 12 h,the chromatographic abundance of E-p-coumaric acid(P6)was higher than that of other metabolites.This phenomenon indicated that E-pcoumaric acid was eliminated in vivo,mainly in its original form within the 12 h period.Between 12 and 24 h,M25 showed a higher detection response than the other compounds in terms of mass spectrometry.M25 was considered as the main form of E-p-coumaricacid eliminated in vivo between 12 and 24 h. The hydrolyzedmetabolite of scopoletin (M5) was present for 24 h, and its glucuronic acid and sulfated conjugates (M12 and M18) were present between 0 and 8 h. In addition, the metabolite produced by thedemethylation and glucuronidation of scopoletin (M6) in vivo wasdetected between 0 and 8 h. Furthermore, two other metabolites ofscopoletin (M7 and M15) were identified in vivo; M7 was consideredto be a demethylation metabolite of scopoletin and M15 was acompound formed by the hydrolysis reaction and sulfation of scopoletin. These two metabolites were present between 0-2 h and4-8 h, and exhibited the phenomenon of “appearing-disappearingreappearing-disappearing” in vivo (Fig. S11). We speculated that themetabolites were secreted into bile and then into the intestinalcavity, with some of the metabolites also excreted with urine. Owing to this, some of these metabolites could then be entered back intoblood circulation via the liver through small intestinal epithelial cells.
Nonetheless,16 metabolites(M8,M9,M10,M16,M19,M20,M24,M27,M28,M29,M35,M51,M57,M60,M64,and M65)of prototype components were not detected in rat urine.For example,a prototype form of ferulic acid was completely absent from the investigated urine samples.Ferulic acid underwent demethylation and existed as caffeic acid(M8).In addition,ferulic acid was further metabolized into ferulic acid-4-O-glucuronic acid(M9)and ferulic acid-4-O-sulfation(M19).It has been reported that M9 and M19 are transformed from ferulic acid in the intestinal epithelium[40].Interestingly,two other metabolites(M16,decarboxylation and sulfation;and M27,decarboxylation and glucuronidation)of ferulic acid that underwent phase I and II reactions in vivo were further characterized.
4.Conclusions
In this study,we developed a post-targeted screening strategy for the analysis of QYSLD-related xenobiotics in rat urine using UPLC-QTOF-MSE.Consequently,20 prototype components and 65 metabolites of QYSLD constituents were characterized.In terms of the metabolites,the constituents in QYSLD mainly underwent oxidation,reduction,sulfation,decarboxylation,hydrolysis,deglycosylation,methylation,glucuronidation,demethylation,acetylation,and binding during different metabolic reactions in vivo.Of the 85 QYSLD-related xenobiotics,14 prototype compounds and 58 metabolites were slowly eliminated and,thus accumulated in vivo over a long period(24 h);five prototypes and two metabolites were present in vivo for a short duration;one prototype and five metabolites underwent the process of“appearing-disappearingreappearing”in vivo.Overall,these findings provide a foundation for further elucidation of the therapeutic mechanism and pharmacokinetics of QYSLD.
CRediT author statement
Ting Zheng:Conceptualization,Methodology,Investigation,Writing-Original draft preparation;Yue Zhao:Formal analysis;Ruijuan Li:Formal analysis;Mengwen Huang:Investigation;An Zhou:Visualization;Zegeng Li:Supervision;Huan Wu:Conceptualization,Writing-Reviewing and Editing,Methodology,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China(Grant No.:81903765),the Open Fund Project of Key Laboratory of Traditional Chinese Medicine for Prevention and Treatment of Major Pulmonary Diseases of the Anhui Provincial Department of Education(Grant No.:JYTKF2020-5),and the Graduate Science and Technology Innovation Fund project of Anhui University of Chinese Medicine(Grant No.:2020YB06).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.05.005.
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Potential therapeutic effects and applications of Eucommiae Folium in secondary hypertension
- Screening of immune cell activators from Astragali Radix using a comprehensive two-dimensional NK-92MI cell membrane chromatography/C18column/time-of-flight mass spectrometry system
- Evaluation of the chemical profile from four germplasms sources of Pruni Semen using UHPLC-LTQ-Orbitrap-MS and multivariate analyses
- Fluorescent intracellular imaging of reactive oxygen species and pH levels moderated by a hydrogenase mimic in living cells
- Sensitive detection of microRNAs using polyadenine-mediated fluorescent spherical nucleic acids and a microfluidic electrokinetic signal amplification chip
- Visualizing the spatial distribution and alteration of metabolites in continuously cropped Salvia miltiorrhiza Bge using MALDI-MSI
