Molecular phylogeny and taxonomy of four Remanella species (Protozoa, Ciliophora): A flagship genus of karyorelictean ciliates, with descriptions of two new species
2022-10-17MingZhenMaYuJieLiuYuanXuBoRongLuYuQingLiSalehALFarrajGiulioPetroniWeiBoSongYingYan
Ming-Zhen Ma, Yu-Jie Liu, Yuan Xu, Bo-Rong Lu, Yu-Qing Li, Saleh A. AL-Farraj, Giulio Petroni, Wei-Bo Song,5,Ying Yan,*
1 Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao, Shandong 266003, China
2 State Key Laboratory of Estuarine and Coastal Research, East China Normal University, Shanghai 200241, China
3 Zoology Department, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
4 Department of Biological, University of Pisa, Pisa 56100, Italy
5 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, Shandong 266237, China
ABSTRACT During faunal studies of psammophilic ciliates along the coast of Qingdao, China, several marine karyorelictean species were isolated. Among them,four species within the genus Remanella were investigated, including two species new to science:i.e., R. rugosa, Remanella elongata sp. nov.,Remanella aposinica sp. nov., and R.unicorpusculata. Remanella rugosa has been reported several times, but this study is the first to provide detailed morphological characters and phylogenetics. Remanella elongata sp. nov. can be distinguished from its congeners by the presence of complex cortical granules, fewer macronuclei, and longer body size. Remanella aposinica sp. nov.differs from its congeners by having 14-17 right lateral ciliary rows and 24-37 dikinetids of intrabuccal kinety. Poorly known Remanella rugosa var. unicorpusculata (Kahl, 1933) Foissner, 1996 should be elevated from subspecies to species level,Remanella unicorpusculata (Foissner, 1996) stat.nov., based on detailed redescriptions with statistical data, living morphology, infraciliature, and species definitions. Small subunit (SSU) rDNA was sequenced for the four species, and phylogenetic analysis revealed that all known taxa in Remanella formed the outline branch to the genus Loxodes with moderate to high bootstrap support among Remanella lineages.
Keywords: Marine ciliates; Morphology;Infraciliature; Phylogeny
lNTRODUCTlON
The enigmatic ciliate class Karyorelictea Corliss, 1974 is widely considered to represent the ancestral ciliate stage (Hu et al., 2019; Lynn, 2008; Song et al., 2009; Wang et al.,2019a; Xu et al., 2013a, 2013b; Yan et al., 2013, 2015, 2016,2017, 2019) due to various unique features, i.e., numerous non-dividing macronuclei (arising from extra divisions of micronuclei) and somatic ciliation with dikinetids (possessing postciliodesmata). Most karyorelictean ciliates live in marine habitats, typically intertidal sandy beaches, and are a key component of benthic energy transfer (Fenchel & Finlay, 1986;Foissner, 1998; Lynn, 2008; Wang et al., 2019b, 2020).Among marine karyorelicteans,Remanellacan be well recognized due to its distinct characteristics such as beak-like anterior rostrum and Müller vesicles. To date, 14 nominal species have been assigned toRemanella(Xu et al., 2013a),eight of which have not been studied by modern morphological techniques (i.e., silver staining, electron microscopy) or molecular methods, resulting in poor information on ciliary patterns, ultrastructures, morphometrics,and molecular data (Foissner, 1996; Xu et al., 2012, 2013a).Therefore, further research onRemanellais required. As more molecular data become available for karyorelicteans and more integrative morphological studies have been performed (e.g.,using living observations, silver staining, and scanning electron microscopy), the inter- and intra-generic relationships within Karyorelictea can be better established (Andreoli et al.,2009; Foissner, 1996; Gao et al., 2010; Mazei et al., 2009; Xu et al., 2012).
In the present study, we report on fourRemanellaspecies isolated from the intertidal zone of a sandy beach on the coast of Qingdao, China.Remanellaelongatasp. nov. andRemanellaaposinicasp. nov. are identified and described as new species. Furthermore,Remanella rugosa(Kahl, 1933)Foissner, 1996 is presented in detail andRemanella unicorpusculata(Kahl, 1933) stat. nov. is redescribed for the first time, providing details on infraciliature for both species.The molecular phylogeny of the four species was analyzed based on newly sequenced small subunit (SSU) rDNA.
MATERlALS AND METHODS
Sample collection, observation, and identification
During ebb tide, ciliates were sampled and isolated from sandy sediment in the intertidal zone of a beach along the coast of Qingdao, China (Figures 1). A 2 cm-3 cm trough was dug, and the top 5 cm of sand and seawater was collected after the trough filled with seawater. Finally, about 10 kg of this mixture was loaded into a bucket. To enrich ciliates in the lab,we used nylon gauze (80 μm-90 μm pore) to filter the collected samples. Firstly, nylon gauze was tied to one end of a polyvinyl plastic tub (50 mm diameter, 100 mm long). About 30 mm of sand was placed in the lower layer of the tube and 30 mm of ice was placed on the upper layer. Seawater and melted freshwater drove the ciliates out of the sand. The filtered samples were collected in Petri dishes (60 mm diameter) (Ma et al., 2021a).
Cells were isolated and observedin vivousing bright field and differential interference microscopy (100× to 1 000×magnifications) with an Olympus BX 53 light microscope(Japan). The infraciliature was revealed using the protargol staining method (Wilbert, 1975). Counts and measurements of impregnated specimens were performed at a magnification of 1 000×. Terminology and systematics primarily followed Foissner (1996) and Lynn (2008), respectively.
DNA extraction, gene amplification, and sequencing
For each species, a single cell was washed with filtered and autoclaved marine water to exclude contamination. Genomic DNA was extracted using a DNeasy Blood & Tissue Kit(Qiagen, Germany) according to the manufacturer’s protocol(Ma et al., 2021a, 2021b). Primers 82F (5'-GAAAC TGCGAATGGCTC-3') (Jerome et al., 1996) and 18S-R (5'-TGATCCTTCTGCAGGTTCACCTAC-3') (Medlin et al., 1988)were used to amplify SSU rDNA by polymerase chain reaction(PCR). To reduce experimental error caused by PCR, Q5®Hot Start High-Fidelity DNA Polymerase (New England Biolabs, USA) was used to amplify the SSU rDNA. The touchdown PCR procedure was performed as follows: one cycle of initial denaturation at 98 °C for 30 s, followed by 18 cycles of amplification (98 °C, 10 s; 69-52 °C, touchdown,30 s; 72 °C, 1 min) and another 18 cycles (98 °C, 10 s; 51 °C,30 s; 72 °C, 1 min), with a final extension of 72 °C for 5 min.The PCR products were purified using an EasyPure Quick Gel Extraction Kit (Transgen Biotech, China), and then cloned using a pClone007 Blunt Simple Vector Kit (Tsingke Biological Technology, China). One clone was randomly selected and cultured in Luria-Bertani (LB) Broth medium for 12 h, then sequenced in two directions by the Tsingke Biological Technology Company (China). The procedure was performed according to Xu et al.(2013a).
Phylogenetic analyses
In addition to the newly characterized SSU rDNA sequences,72 sequences were obtained from the NCBI GenBank database. Five heterotrichs were selected as outgroup species. Sequence alignment was carried out using the GUIDANCE algorithm (Penn et al., 2010) with default parameters on the GUIDANCE web server (Penn et al., 2010).The resulting alignment was manually edited using BioEdit v7.0.5.2 (Hall, 1999). The final alignment, including 1 694 sites and 76 taxa, was used to construct phylogenetic trees.
Bayesian inference (BI) analysis was performed with MrBayes v3.1.2 (Ronquist et al., 2012) using the GTR+I+G model as selected by Akaike information criterion (AIC) in MrModeltest v.2.0 (Nylander, 2004). The BI analysis was conducted with 1 000 000 generations and sampling every 100 generations. The first 25% of sampled trees were discarded as burn-in. Maximum-likelihood (ML) analysis was carried out with 1 000 replicates using the CIPRES Portal v2.0 (http://www.phylo.org) and RAxML-HPC2 on XSEDE with the GTRGAMMA model (Stamatakis et al.,2008). MEGA v.5.0(Tamura et al., 2011) was used to visualize phylogenetic tree topology.
Topology testing

Figure 1 Sample sites
The statistical possibility of alternative phylogenetic hypotheses was evaluated using approximately unbiased (AU)tests (Shimodaira, 2002) to assess phylogenetic relationships among different taxa within Karyorelictea. Two constrained ML trees were generated using RAxML v8.2.10 (Stamatakis,2014) with enforced constraints and then compared with unconstrained ML topologies implemented in CONSEL(Shimodaira & Hasegawa, 2001).
RESULTS
Class Karyorelictea Corliss, 1974 Family Loxodidae Bütschli, 1889 Genus Remanella Foissner, 1996
Remanella rugosa (Kahl, 1933) Foissner, 1996 (Figures 2,3; Table 1)
This species was first reported by Kahl (1933) based on observations of its living morphology. Several researchers later provided sketches of living species (Carey & Maeda,1985; Dragesco, 1960; Kattar, 1970). Raikov (1993) described its extrusomes and Foissner (1996) depicted its infraciliature but without detailed description. Therefore, a redescription with observation on both living and stained specimens is needed. Here, we provide an improved diagnosis based on previous and present observations and a detailed redescription based on the Chinese population.
lmproved diagnosis:Cell sizein vivo90-400 μm×20-50 μm;ratio of buccal field/body length about 1/5-1/3; 11-15 right lateral ciliary rows; right buccal, left outer buccal, and intrabuccal kinety composed of 40-60, 14-25, and 23-40 dikinetids, respectively; 2-3 macronuclei; single micronucleus;cortical granules brown; 3-10 Müller vesicles; marine habitat.Voucher material:A voucher slide of protargol-impregnated specimens was deposited in the Laboratory of Protozoology,Ocean University of China (OUC), Qingdao, China(Registration No. MMZ2020060202).
Description:Live cells slender and rather flexible, 90-200 μm×20-50 μm; most about 110 μm×35 μm in size, with buccal field occupying ca. 1/5 of body length (Figure 2A-C;Figure 3A, B). Cilia about 6 μm long. Cells mostly dark brown at low magnification due to brown cortical granules. Cortical granules ca. 0.5 μm in diameter and densely packed in buccal area, around pharyngeal tube, and on both cell sides (Figure 2F,G; Figure 3C-E). On right side of cell, cortical granules arranged in lines along ciliary rows (Figure 2G; Figure 3C, E).On left side of cell, cortical granules scattered (Figure 2F;Figure 3D). Five to ten Müller vesicles located near dorsal margin of cell, each 7 μm in diameter with a globular or ellipsoidal mineral granule ca. 2 μm×5 μm across (Figure 2A-C; Figure 3A, B). Cytoplasmic spicules about 10 μm long,scattered throughout cell (Figure 2A, G; Figure 3A, B, F, H).Many refractive globular granules scattered throughout cell(Figure 3G, H). Two to three macronuclei with single closely associated micronucleus (Figure 2D; Figure 3F, J, K).Locomotion by gliding between sand grains or along bottom of Petri dish.

Figure 2 Remanella rugosa (Kahl, 1933) Foissner, 1996 from life (A-G) and after protargol staining (H-K)
Entire infraciliature consisting of dikinetids. Right surface densely ciliated with 12-14 right lateral ciliary rows (Figure 2H;Figure 3I, L). Dorsolateral kinety extending to posterior end of cell (Figure 2H; Figure 3I). Left lateral ciliary row extending around entire cell margin (Figure 2I). Right buccal kinety longitudinal along right margin of buccal overture, composed of 40-60 tightly spaced dikinetids (Figure 2J, K; Figure 3M).Left outer buccal kinety longitudinal along left margin of buccal overture, composed of 17-25 closely spaced dikinetids(Figure 2J, K). Intrabuccal kinety consisting of 23-40 dikinetids (Figure 2J, K).
Remanella elongata sp. nov. (Figures 4, 5; Table 1)
Diagnosis:Cell sizein vivo250-500 μm×50-70 μm; buccal field occupying ca. 1/4-1/3 of body length; 18-22 right lateral ciliary rows; right buccal, left outer buccal, and intrabuccal kinety composed of 125-180, 50-100, and 105-150 dikinetids, respectively; 1-3 macronuclei, single micronucleus;cortical granules tiny, brown, in lines distributed in both sides of glabrous stripe and along ciliary rows, but sparsely in middle of glabrous stripe; 7-13 Müller vesicles. Marine habitat.
Type locality, ecological features, and sample date:Intertidal zone of Silver Beach at Qingdao (N35°55′09″,E120°11′55″), China (Figure 1). Water temperature was 24 °C and salinity was ca. 30‰. Sample was collected on 13 June 2020.
Type specimens:A protargol-impregnated slide containing the holotype specimen marked with an ink circle was deposited in the Laboratory of Protozoology, OUC, China (No.MMZ2020061304).
Etymology:The species group name “elongata” refers to the slender body shape of the species.

Figure 3 Photomicrographs of Remanella rugosa (Kahl, 1933) Foissner, 1996 from life (A-H) and after protargol staining (l-O)
Description:Live cells 250-500 μm×50-70 μm; most about 400 μm×60 μm in size, with buccal field occupying ca. 1/4-1/3 of body length (Figure 4A-D; Figure 5A-E). Cilia about 10 μm long. Cells brown at low magnification (×50) due to many rows of highly refractile cortical granules. Cells appearing dark at mid-body, with transparent anterior and tail at low magnification due to presence or absence of multiple refractile particles. Cortical granules tiny (ca. 0.5 μm in diameter),brown, in lines on both sides of glabrous stripe. Longitudinal bright “line” appearing due to sparsely scattered cortical granules in middle of glabrous stripe. Cortical granules densely covering margin in left lateral region of cell (Figure 4E,G; Figure 5G, H). Approximately 7-13 Müller vesicles for gravity reception, 10 μm in diameter, with globular or ellipsoidal mineral content ca. 5 μm in diameter, located near dorsal margin, mostly gathering at anterior; several small developing Müller vesicles dispersed in cell (Figure 4A-D, I;Figure 5A-F). Spicules forming unique cytoskeleton, most about 15 μm long, scattered throughout cell (Figure 4A, I;Figure 5F, L). Food vacuole observed with diatom inside(Figure 5I), suggesting consumption of diatoms. Glides through sand grains and on bottom of Petri dish, sometimes undulating.
Infraciliature consisting of dikinetids. Right surface densely ciliated, gradually shortening anteriorly at posterodorsal margin of cell, especially posteriorly at ventral margin, with 18-22 right lateral ciliary rows (Figure 4J; Figure 5P).Dorsolateral kinety extending to end of tail (Figure 4J). Left side barren. Left lateral ciliary row curving around cell(Figure 4K; Figure 5K). Right buccal kinety longitudinal along right margin of buccal overture, composed of 125-180 tightly spaced dikinetids (Figure 4H; Figure 5J, O). Left outer buccal kinety longitudinal along left margin of buccal overture,composed of 50-100 closely spaced dikinetids (Figure 4H;Figure 5J, N). Intrabuccal kinety consisting of 105-150 dikinetids (Figure 4H; Figure 5J, O). One to three macronuclei with single micronucleus in-between (Figure 4F; Figure 5L, M).
Remanella aposinica sp. nov. (Figures 6, 7; Table 1)
Diagnosis:Cell sizein vivoabout 160-285 μm×35-55 μm;buccal field occupying ca. 1/6-1/5 of body length; 14-17 right lateral ciliary rows; right buccal, left outer buccal, and intrabuccal kinety composed of 75-105, 15-25, and 24-37 dikinetids, respectively; 2-3 macronuclei; single micronucleus;brown cortical granules; single Müller vesicle. Marine habitat.
Type locality, ecological features, and sample date:Intertidal zone of First Bathing Beach at Qingdao (N36°03′24″,E120°20′32″), China (Figure 1). Water temperature was 20 °C and salinity was ca. 30‰. Sample was collected on 6 March 2019.
Type specimens:A protargol-impregnated slide containing the holotype specimen marked with an ink circle was deposited in the Laboratory of Protozoology, OUC, China (No.MMZ2019030601).
Etymology:The epithet is composed of the Greek prefix αϖó(apo, derived from) and species group namesinica, reflecting the superficial similarity of this species toR. sinicaXu et al.,2012.

Table 1 Morphometric data for Remanella rugosa (R. rug), Remanella elongata sp. nov. (R. elo), Remanella aposinica sp. nov. (R. apo),and Remanella unicorpusculata (Foissner, 1996) stat. nov. (R. uni)
Description:Live cells 160-285 μm×35-55 μm, most about 200 μm×35 μm in size, with buccal field occupying ca. 1/6-1/5 of body length (Figure 6A, B; Figure 7A, B), narrow due to flattening organism. Tail short and inconspicuous (Figure 7E).Cilia about 10 μm long. Cells rather dark, almost brown due to brown cortical granules ca. 0.5 μm in diameter and densely packed in buccal area, around pharyngeal tube, and on both cell sides except mid-region of left side (Figure 6G-I;Figure 7D, G). Cortical granules conspicuously absent from longitudinally oriented “groove” formed by many transverse folds on left side, appearing as bright “line” when observedin vivo(Figure 6I; Figure 7G).On right side of cell, granules arranged in lines along ciliary rows (Figure 6H). Single Müller vesicle about 8 μm in diameter, with globular mineral granule inside, located near dorsal margin of cell, level with posterior end of buccal area (Figure 6A-C; Figure 7A-C). Cytoplasmic spicules about 10 μm long, scattered throughout cell(Figure 7K, N). Alga observed in body (Figure 7F). Cell surface of left side rather rough due to many transverse folds(Figure 6I; Figure 7G). Two or three macronuclei with single closely associated micronucleus (Figure 6D; Figure 7H, I).Locomotion by gliding between sand grains or along bottom of Petri dish.
Entire infraciliature consisting of dikinetids. Left lateral ciliary row extending around entire cell margin (Figure 6K;Figure 7K); 14-17 right lateral ciliary rows (Figure 6J;Figure 7O); dorsolateral kinety extending to posterior end of cell (Figure 6J; Figure 7L). Right buccal kinety longitudinal along right margin of buccal overture, composed of 75-105 tightly spaced dikinetids. Left outer buccal kinety longitudinal along left margin of buccal overture, composed of 15-25 closely spaced dikinetids. Intrabuccal kinety composed of 24-37 dikinetids (Figure 6E, F; Figure 7J, K, M).

Figure 4 Remanella elongata sp. nov. from life (A-G, l) and after protargol staining (H, J-K)
Remanella unicorpusculata (Kahl, 1933) stat. nov.Synonyms: Remanella unicorpusculata Dragesco, 1965 Remanella rugosa var. aunicorpusculata (Kahl, 1933)Foissner, 1996 (Figures 8, 9; Table 1)
This species was first reported by Kahl (1933), who provided a brief sketch and description of a single-Müllervesicle variety ofRemanella rugosa, namedRemanella rugosavar.unicorpusculata. Later, Dragesco (1965) named it asRemanella unicorpusculata, with a description of body length, number of right lateral ciliary rows, and number of macronuclei.However, no information on its oral ciliature has been given. In addition, the genusRemanellawas not valid untilR. multinucleata(Kahl, 1933) Foissner, 1996 was fixed as the type species (Foissner, 1996). According to Foissner(1996),Remanella unicorpusculatawas an invalid species due to a misinterpretation of the International Commission on Zoological Nomenclature (ICZN) in Dragesco (1965).Nevertheless, the established species from Dragesco (1965)can be clearly distinguished fromR. rugosaby its single Müller vesicle and smaller body size. Therefore, in the present study,we elevateRemanella rugosavar.unicorpusculatatoRemanella unicorpusculata(Kahl, 1933)stat. nov.and provide an improved diagnosis and a redescription based on present and previous studies.
lmproved diagnosis:Cell sizein vivo90-140 μm×15-20 μm;
buccal field occupying ca. 15%-25% of body length; 7-10 right lateral ciliary rows; right buccal, left outer buccal, and intrabuccal kinety composed of 20-33, 3-5, and 9-20 dikinetids, respectively; 2-3 macronuclei; single micronucleus;brown cortical granules; one Müller vesicle. Marine habitat.
Voucher slides:Voucher slides of protargol-impregnated specimens from Qingdao population were deposited in the Laboratory of Protozoology, OUC, China (Registration No.:MMZ2020060801).
Type locality, ecological features, and sample date:
Intertidal zone of First Bathing Beach at Qingdao (N36°03′24″,E120°20′32″), China (Figure 1). Water temperature was 24 °C and salinity was ca. 30‰. Sample was collected on 8 June 2020.
Description of population from China:Live cells 90-140 μm×15-20 μm, most about 110 μm×18 μm in size, with buccal field occupying ca. 15%-25% of body length (Figure 8A;Figure 9A-D). Cilia about 10 μm long. Cells rather transparent and almost colorless. Cell surface of left side rather rough,brown cortical granules sparsely scattered (Figure 8D;Figure 9E). One Müller vesicle, about 6 μm-8 μm in diameter with ellipsoidal mineral content, ca. 3 μm-5 μm in diameter,located near dorsal margin in anterior portion; 1-2 developing Müller vesicles present in middle region of posterior portion(Figure 8A-C; Figure 9A, B, F). Cytoplasmic spicules about 10 μm-15 μm long, scattered throughout cell. Alga observed in body (Figure 9H, I). Two or three macronuclei with single micronucleus in-between (Figure 8E; Figure 9G, N, O).
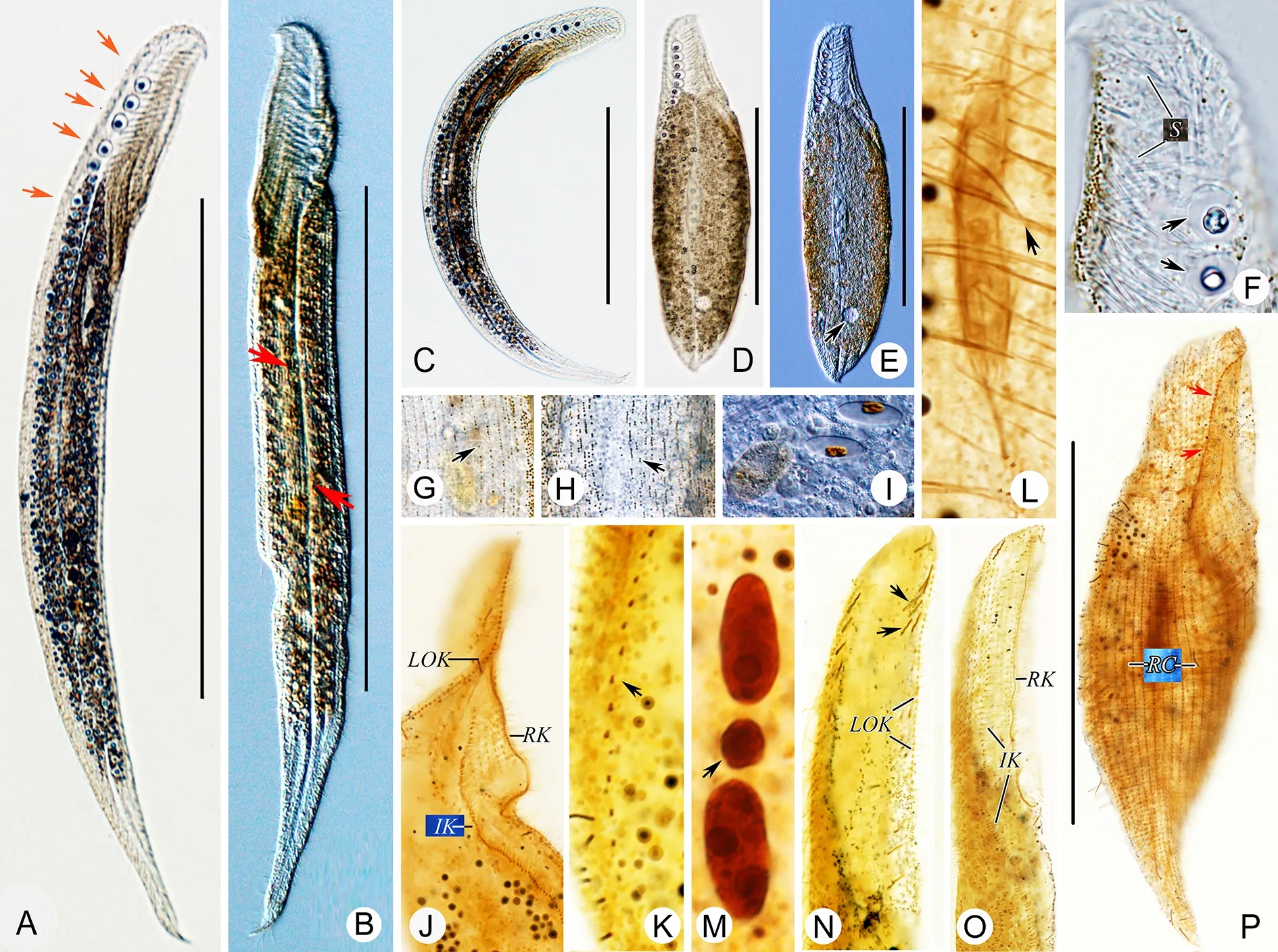
Figure 5 Remanella elongata sp. nov. from life (A-l) and after protargol staining (J-P)
Infraciliature consisting of dikinetids. Right surface densely ciliated, with 7-10 right lateral ciliary rows (Figure 8H;Figure 9P). Dorsolateral kinety extending to end of tail(Figure 8G; Figure 9M). Left lateral ciliary row curving around cell (Figure 8I; Figure 9Q). Right buccal kinety longitudinal along right margin of buccal overture, composed of 20-33 tightly spaced dikinetids (Figure 8F, H; Figure 9L, P). Left outer buccal kinety composed of 3-5 dikinetids. Intrabuccal kinety consisting of 9-20 dikinetids (Figure 8F, H, I;Figure 9J-L).
Molecular data and phylogenetic analyses
Sequence information:The sequence lengths ofRemanella rugosa,Remanella elongatesp. nov.,Remanella aposinicasp. nov., andRemanella unicorpusculata(Kahl, 1933)stat.nov.were 1 546, 1 547, 1 545, and 1 546 bp, respectively. The GC contents were 47.54%, 48.09%, 47.96%, and 47.80%,respectively. GenBank accession Nos. of the four newly generated sequences were OM127343, OM127344,OM127345, and OM127342, respectively.
Phylogenies inferred from SSU rDNA:The ML and BI trees showed similar topologies and thus only the ML tree topology was presented (Figure 10). The family Loxodidae was wellsupported as a monophyletic group (97% ML, 1.00 BI),forming a sister clade to the family Trachelocercidae (69% ML,1.00 BI).Remanellaformed a well-supported clade basal to the genusLoxodesin phylogenetic analyses.The hypothesis of the genusRemanellabeing monophyletic was rejected based on the AU test (P=0.017<0.05). The four newly sequencedRemanellaspecies were all contained within Loxodidae, as expected.Remanellaelongatasp. nov.grouped withR. caudata, together clustering with theLoxodesspecies.The sequences ofR. rugosaandR.sp. (JX015378)were identical, and thus they clustered together with full support.Remanellaelongatesp. nov.andR. caudatagrouped together with weak support, with a 71 bp difference between in their sequences.Remanellaunicorpusculatastat.nov. branched withRemanellasp. (JX015377), with a 182 bp difference between their sequences (Supplementary Table S1).Remanella aposinicasp. nov. clustered withR. sinicawith strong support (94% ML, 1.00 BI) and a 6 bp difference,then together grouped withRemanellasp. (AM409181),forming an early-branching lineage with Loxodidae.
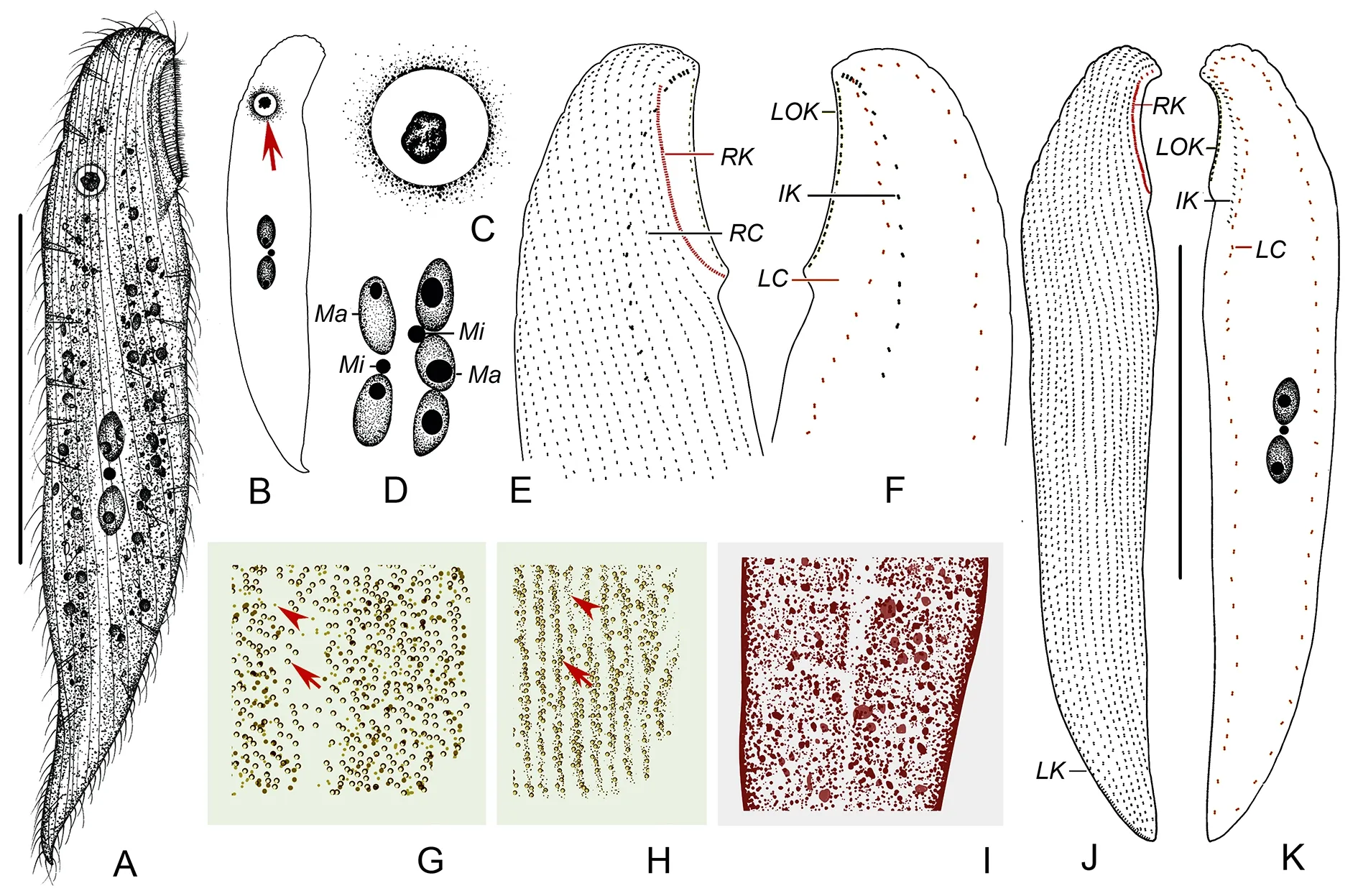
Figure 6 Remanella aposinica sp. nov. from life (A-D, G-l) and after protargol staining (E-F, J-K)
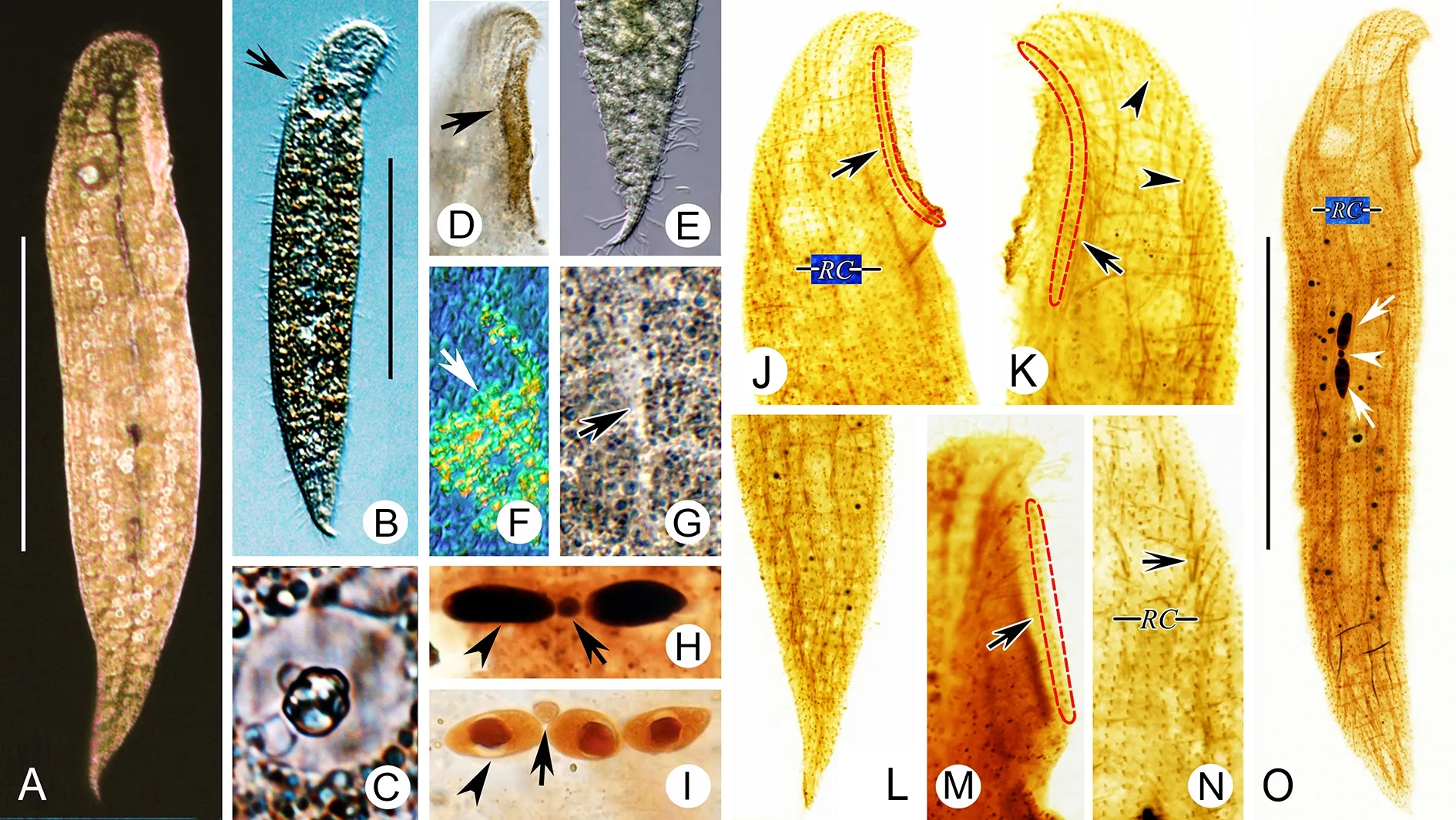
Figure 7 Remanella aposinica sp. nov. from life (A-G) and after protargol staining (H-O)

Figure 8 Remanella unicorpusculata (Kahl, 1933) stat. nov. from life (A-E) and after protargol staining (F-l)

Figure 9 Remanella unicorpusculata (Kahl, 1933) stat. nov. from life (A-l) and after protargol staining (J-Q)
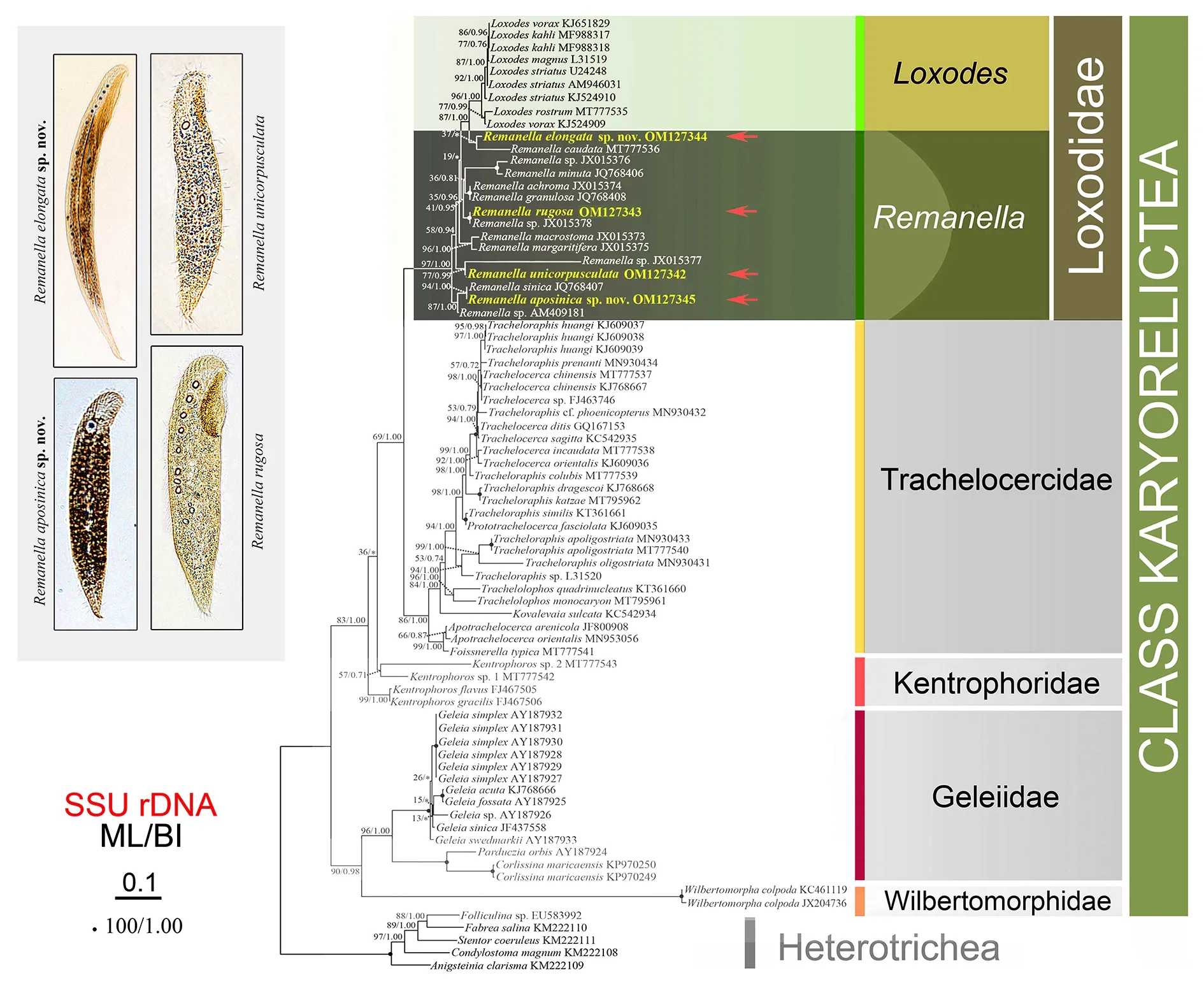
Figure 10 ML tree inferred from SSU rDNA sequences showing phylogenetic positions of four newly sequenced species
DlSCUSSlON
Comments on Remanella rugosa (kahl, 1933) Foissner,1996
Remanella rugosawas originally described by Kahl (1933) as“Size 200-300 μm, two elongated nuclei…Müller vesicles 3 to 8 … ” (translated from German). Considering the general morphology, the Chinese population corresponds well with the original description (Figure 2E). The main difference between the two populations is body size (200 μm-300 μm in the original population vs. usually 90 μm-200 μm in the Chinese population). However, this difference may be due to its contractility. Therefore, the Chinese population is recognized as a new population ofR. rugosa.
The species was later reported with more or fewer details in many places (Carey & Maeda, 1985; Dragesco, 1960;Hartwig, 1973; Kattar, 1970; Lepsi, 1962; Raikov, 1993).However, its infraciliature was not reported until Foissner(1996) but only by means of a figure and without a full redescription or morphometric data. Most other populations are similar to the Chinese population, except for the following.The population described by Dragesco (1960) has a larger body size range (180 μm-400 μm vs. 90 μm-200 μm) and higher number of right lateral ciliary rows than the Chinese population (14-15 vs. 12-14). The sketch in Foissner (1996)also shows one left inner buccal kinety near the left outer buccal kinety. However, as there is no additional description of the sketch, the identification of this population should be further considered.
Remanella rugosais similar toR. brunneabut differs by buccal to body length ratio (1/5-1/3 inR. rugosavs. 1/9-1/5 inR. brunnea) (Dragesco, 1965).

Figure 11 lllustration of updated identification key of 16 current species within Remanella
Comments on Remanella elongata sp. nov.
Considering the general morphology, including body shape,number of macronuclei, right lateral kineties, right buccal, left outer buccal, intrabuccal kinety, and color of cortical granules,Remanella granulosa(Kahl, 1933) Foissner, 1996 andRemanella multinucleata(Kahl, 1933) Foissner, 1996 should be compared withRemanella elongatasp. nov.(Figure 11 and Table 2).
Remanella granulosadiffers fromRemanella elongatasp.nov.by having sparsely scattered cortical granules in glabrous stripe (Xu et al., 2012) vs. cortical granules arranged in lines on both sides of glabrous stripe, sparse in middle of glabrous stripe, appearing as longitudinal bright “line” when observedin vivo(Figure 4G; Figure 5H).
Remanella multinucleatacan be distinguished fromRemanella elongatasp. nov.by longer body size (500 μm-1 000 μm vs. 250 μm-500 μmin vivo), different color and arrangement of cortical granules (yellowish, in rows vs. brown,complex arrangement), and more macronuclei (7-24 vs. 1-3).
Comments on Remanella aposinica sp. nov.
Remanella aposinicasp. nov.is mainly characterized by having one Müller vesicle. Considering this feature,R. sinicaXu et al., 2012,R. unicorpusculataFoissner, 1996,R.dragescoi(Agamaliev, 1966) Foissner, 1996, andR.microstoma(Dragesco, 1954) Foissner, 1996 should be compared with the new species (Figure 11).Remanella sinicais similar toRemanella aposinicasp. nov.in general morphologyin vivoand by having a single Müller vesicle(Table 2) but can be distinguished by having more right lateral ciliary rows (18-20 vs. 14-17). In addition, their SSU rDNA sequences differ by 6 bp, indicating that these two organisms are not conspecific (Xu et al., 2012).
Remanellaunicorpusculatadiffers fromRemanella aposinicasp. nov.by its smaller size (90 μm-140 μm vs. 160 μm-285 μm) and fewer right lateral ciliary rows (7-10 vs.14-17) (Dragesco, 1965; Table 2).
There is no information on the infraciliature ofR. dragescoi.However, from the original report, it can be inferred thatR.dragescoidiffers fromRemanella aposinicasp. nov.by having more macronuclei (6 vs. 2-3) (Agamaliev, 1966).
Information on the infraciliature ofR. microstomais also lacking. Nevertheless, this species can be easily distinguished fromRemanella aposinicasp. nov.by its distinct and narrow tail (ca. 1/4 of body length vs. short and inconspicuous),smaller body size (140 μm vs. 160 μm-285 μm), and smaller buccal field/body length ratio (1/7 vs. 1/6-1/5) (Dragesco,1954b).
Comments on Remanella unicorpusculata (Kahl, 1933)stat. nov.
Remanella unicorpusculata(Kahl, 1933)stat. nov.is characterized by one Müller vesicle and smaller body size.According to Xu et al.(2013a), the number of Müller vesicles is an important character for identifying species in Loxodidae.Considering the differences in Müller vesicle numbers betweenR. rugosa unicorpusculata(Kahl, 1933) Foissner,1996 andR. rugosa(1 vs. 3-8), we agree with Dragesco(1965) on elevating this species to species-rank,Remanella unicorpusculata(Kahl, 1933)stat. nov.(Table 2).Furthermore, our population corresponds well with the original description and that of Dragesco (1965), i.e., “85-100 μm long, one Müller vesicles, ten right lateral ciliary rows, brown cortical granules, two macronuclei and one micronucleus…”.
Similar to the comparisons forRemanella aposinicasp.nov. classification,Remanella unicorpusculata(Kahl, 1933)stat. nov. should also be compared withR. sinicaXu et al.,2012,R. dragescoi(Agamaliev, 1966) Foissner, 1996, andR.microstoma(Dragesco, 1954) Foissner, 1996, which are all characterized by single Müller vesicles.
Remanella sinicadiffers fromRemanella unicorpusculata(Kahl, 1933) stat. nov. based on its larger size (200 μm-320 μm vs. 90 μm-140 μm) and more right lateral ciliary rows(18-20 vs. 7-10) (Xu et al., 2012).
From its original description,R. dragescoidiffers fromRemanella unicorpusculata(Kahl, 1933) stat. nov. by having more macronuclei (6 vs. 2-3) (Agamaliev, 1966).
With its distinct features,R. microstomacan be distinguished fromRemanella unicorpusculata(Kahl, 1933)stat. nov. based on its narrow tail (ca. 1/4 of body length vs.short and wedge-shaped (Dragesco, 1954b).
Comments on phylogenetic analyses
The class Karyorelictea is a stable monophyletic group, as reported previously (Ma et al., 2022; Xu et al., 2012, 2013a).The class can be subdivided into five families: Loxodidae,Geleiidae, Kentrophoridae, Wilbertomorphidae, and Trachelocercidae. Loxodidae and Trachelocercidae have a close relationship. Within Loxodidae,Loxodesis located withinRemanella, well reflecting previous reports that freshwaterLoxodesis likely to have evolved from marineRemanella(Xu et al., 2013a).Remanella unicorpusculata,R. sinica,R.aposinica, and the other twoRemanellasp. form the two early branches. In addition, single Müller vesicles may be an ancestral character.
ldentification key of genus Remanella
Based on previous studies and our present work, an updated key to the identification of species within the genusRemanellais provided (Figure 11):


NOMENCLATURAL ACTS REGlSTRATlON
The electronic version of this article in portable document format will represent a published work according to the International Commission on Zoological Nomenclature (ICZN),and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone (see Article 8.5-8.6 of the Code). This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for ICZN.The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to prefix http://zoobank.org/.Publication LSID:
urn:lsid:zoobank.org:pub:1ACD9A93-8BB0-454E-91B7-6CCF ECD396C0
Nomenclatural act LSID:
Remanella elongatasp. nov. urn:lsid:zoobank.org:act:62EB 1882-EAF3-4BD5-8516-402903DEAD5D
Remanella aposinicasp. nov. urn:lsid:zoobank.org:act:28E 326F3-6BBC-4AA9-B00E-EF7B68DA45CF
AUTHOR CONTRlBUTlONS
M.Z.M. and Y.J.L. performed the experiments and data analyses. M.Z.M. wrote the original draft. Y.X., B.R.L., Y.Q.L.,S.A.A., G.P., W.B.S., and Y.Y. reviewed and edited the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
Many thanks are due to Dr. Zhe Wang (Shandong University)for his help with topology testing.
杂志排行
Zoological Research的其它文章
- Description of two new species of Hemiphyllodactylus(Reptilia: Gekkonidae) from karst landscapes in Yunnan, China, highlights complex conservation needs
- Functional explanation of extreme hatching asynchrony: Male Manipulation Hypothesis
- Metagenomic analysis reveals presence of different animal viruses in commercial fetal bovine serum and trypsin
- Phylogenetic analysis of combined mitochondrial genome and 32 nuclear genes provides key insights into molecular systematics and historical biogeography of Asian warty newts of the genus Paramesotriton(Caudata: Salamandridae)
- Dwindling in the mountains: Description of a critically endangered and microendemic Onychodactylus species (Amphibia, Hynobiidae) from the Korean Peninsula
- Whole-genome resequencing reveals molecular imprints of anthropogenic and natural selection in wild and domesticated sheep
