Identification of key genes involved in axon regeneration and Wallerian degeneration by weighted gene co-expression network analysis
2022-08-27YanLuQiShanMeiLingXiAnNiSuSuMaoBinYuQianQianCao
Yan Lu ,Qi Shan ,Mei Ling ,Xi-An Ni ,Su-Su Mao,Bin Yu,,Qian-Qian Cao,
Abstract Peripheral nerve injury repair requires a certain degree of cooperation between axon regeneration and Wallerian degeneration.Therefore,investigating how axon regeneration and degeneration work together to repair peripheral nerve injury may uncover the molecular mechanisms and signal cascades underlying peripheral nerve repair and provide potential strategies for improving the low axon regeneration capacity of the central nervous system.In this study,we applied weighted gene co-expression network analysis to identify differentially expressed genes in proximal and distal sciatic nerve segments from rats with sciatic nerve injury.We identified 31 and 15 co-expression modules from the proximal and distal sciatic nerve segments,respectively.Functional enrichment analysis revealed that the differentially expressed genes in proximal modules promoted regeneration,while the differentially expressed genes in distal modules promoted neurodegeneration.Next,we constructed hub gene networks for selected modules and identified a key hub gene,Kif22,which was up-regulated in both nerve segments. In vitro experiments confirmed that Kif22 knockdown inhibited proliferation and migration of Schwann cells by modulating the activity of the extracellular signal-regulated kinase signaling pathway.Collectively,our findings provide a comparative framework of gene modules that are co-expressed in injured proximal and distal sciatic nerve segments,and identify Kif22 as a potential therapeutic target for promoting peripheral nerve injury repair via Schwann cell proliferation and migration.All animal experiments were approved by the Institutional Animal Ethics Committee of Nantong University,China (approval No.S20210322-008) on March 22,2021.
Key Words:axon regeneration;extracellular signal-regulated kinase signaling pathway;hub genes;Kif22;peripheral nerve injury;protein kinase Cα;Schwann cells;Wallerian degeneration;weighted gene co-expression network analysis
Introduction
Peripheral nerve injury often occurs during surgery,and refers to primary or secondary lesions within the peripheral nervous system.Trauma,tumor,metabolic disease,and other factors can induce peripheral nerve injury,leading to sensory,motor,and nutritional disorders in the innervated region.Unlike the central nervous system,the peripheral nervous system has regenerative capacity,which involves an orderly series of morphological and molecular events that lead to repair following injury (He and Jin,2016).Injury that severs a nerve completely results in division of the axon into proximal and distal nerve stumps:the nerve stump connected to the cell body is referred to as the proximal axon segment,and the disconnected nerve stump is referred to as the distal axon segment (Girouard et al.,2018).After peripheral nerve injury,the axon begins to regenerate from the proximal stump,while the distal stump undergoes Wallerian degeneration (WD)(Coleman and Freeman,2010;Girouard et al.,2018).
In recent years,considerable progress has been made in understanding the mechanism of peripheral nerve repair(He and Jin,2016;Mahar and Cavalli,2018).In particular,the intrinsic molecular mechanisms that separately underlie axon regeneration and degeneration are now well understood(Coleman and Freeman,2010;Conforti et al.,2014;Mahar and Cavalli,2018).However,proximal axon regeneration and distal degeneration are not completely independent processes.A previous study showed that successful nerve repair requires a degree of coordination between axon regeneration and WD (Girouard et al.,2018).Degeneration of the distal axon segment creates conditions that induce axon regrowth from proximal stumps,thus influencing the regenerative capacity of the proximal axon segment.In addition,several molecules,such as Dual leucine zipper kinase,c-Jun N-terminal kinase,calpain,stathmin 2,and nicotinamide nucleotide adenylyltransferase 2,and some cellular processes,including Schwann cell (SC) dedifferentiation and proliferation,axonal transport,mitochondrial dynamics,and cytoskeletal dynamics have been shown to play essential roles in both axon regeneration and degeneration (Jessen and Mirsky,2016;Prior et al.,2017;Blanquie and Bradke,2018;Girouard et al.,2018;Smith and Gallo,2018).Therefore,investigating how axon regeneration and degeneration work together to regulate peripheral injury repair may not only reveal the molecular mechanisms and signal cascades involved in peripheral nerve repair,but also suggest potential strategies to improve proximal axon segment regeneration in the central nervous system,which lacks regenerative capacity.
To understand how axon regeneration and degeneration work together to regulate peripheral injury repair,we sought to generate a comprehensive framework of gene expression in proximal and distal nerve segments.Microarray and RNAsequencing,two emerging high-throughput sequencing technologies,were used to systematically monitor gene expression profile changes during axonal regeneration and degeneration,respectively.Previous work from our group used microarray analysis to reveal transcriptome alterations in the proximal and distal segments of sciatic nerves undergoing WD (Yao et al.,2013;Jiang et al.,2014;Yu et al.,2016;Yi et al.,2017).However,these earlier studies did not systematically analyze and compare gene expression profiles between the proximal and distal nerve segments.To address this,in the current study we applied weighted gene co-expression network analysis (WGCNA),an efficient and accurate bioinformatics and biological data mining method,along with a comprehensive collection of R functions that are used to perform various aspects of weighted correlation network analysis,and frequently to analyze interactions between genes and phenotypes (Zhang and Horvath,2005;Liu et al.,2017).WGCNA can help compare differentially expressed genes,as well as identify relationships among genes in different co-expression modules (Liu et al.,2017;Wan et al.,2018).The aim of the current study was to use WGCNA to compare co-expression gene modules between proximal and distal nerve segments and to identify genes that play key roles in regulating peripheral nerve repair.
Kinesin family member 22 (Kif22) encodes a motor protein that transports organelles within cells and moves chromosomes during cell division.Kif22has been previously reported to regulate extracellular signal-regulated kinase(ERK) signal pathway-induced cell proliferation and migration,as well as epidermal growth factor receptor signaling (Yu et al.,2014,2020;Pike et al.,2018).Given that cell division,proliferation,and migration are essential for nerve repair and that the ERK and epidermal growth factor receptor signaling pathways play important roles in nerve repair (Zhou and Li,2007;Napoli et al.,2012),we investigated whetherKif22plays a role in peripheral nerve injury repair.
Materials and Methods
Animal surgery and tissue collection
Changing estrogen levels in female mice would have interfered with analysis of gene expression after nerve injury,so only male mice were used in this study.One hundred and fortyseven adult male Sprague-Dawley rats (specific-pathogenfree level,8-week-old,weighing 160–200 g) were provided by the Experimental Animal Center of Nantong University(license Nos.SYXK (Su) 2014-0001 and SYXK (Su) 2017-0046).All animals were maintained and used in accordance with the guidelines of Institutional Animal Care of Nantong University.All animal experiments were approved by the Institutional Animal Ethics Committee of Nantong University,China(approval No.S20210322-008) on March 22,2021.The rats were randomly divided into seven groups (0-,1-,and 6-hour and 1-,4-,7-,and 14-day groups),each of which included six rats.All of the rats were subjected to sciatic nerve transection surgery,as previously described (Jiang et al.,2014).Briefly,the sciatic nerve was exposed through an incision in the midlateral area of the thigh of the left hind leg,and a 10-mm nerve segment was excised.At 0,1,or 6 hours or 1,4,7,or 14 days post-surgery,the rats were sacrificed by abdominal injection of a mixture of 85 mg/kg trichloroacetaldehyde monohydrate(Lingfeng Chemical Reagent,Shanghai,China),42 mg/kg magnesium sulfate (Lingfeng Chemical Reagent),and 17 mg/kg sodium pentobarbital (Millipore Sigma,Burlington,MA,USA),and 0.5-cm segments of the proximal and distal nerve stumps were harvested.
Microarray analysis
Microarray analysis was carried out as previously described (Yi et al.,2017).Total RNA was extracted from the proximal and distal nerve segments of rats with sciatic nerve injury using Trizol reagent (Sigma,San Francisco,CA,USA) according to the manufacturer’s instructions.RNeasy spin columns (Qiagen,Valencia,CA,USA) were used to remove contaminating DNA.Then,RNA quality assessment,amplification,labeling,hybridization,and further microarray analysis of each sample were conducted by the Shanghai Biotechnology Corporation,Shanghai,China.Three biological replicates were included for each group.
Weighted gene co-expression network analysis
WGCNA was carried out using the WGCNA R software package (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/).Differentially expressed genes identified by microarray analysis were then subjected to co-expression network analysis.First,we filtered out genes with low variation in expression (standard deviation≤ 0.25).Then,we used a Pearson correlation matrix to perform co-expression analysis of paired genes.We selected an adjacency matrix weight parameter power of 14,based on a previous report,to satisfy the preconditions of scalefree network distribution as much as possible.Finally,based on this selected power value,we established a weighted coexpression network model that divided the differentially expressed genes in the proximal and distal groups into 31 and 15 modules,respectively.
Identification of significant modules and key genes
To identify the important modules,a Pearson correlation algorithm was applied to compute the correlation coefficient andPvalue of module characteristic genes and trait (time post-injury).Then,an absolute correlation coefficient value≥ 0.5 and aPvalue <0.05 were used as thresholds to screen for modules related to the trait.Once the modules of interest were selected,the correlation between module gene expression and corresponding traits (gene significance,GS),as well as the correlation between module gene expression and module characteristic gene (module membership,MM)was calculated for each important gene.Then,thresholds of MM >0.9 and GS >0.5 were used to identify key genes in the indicated modules.
Functional enrichment analysis of co-expression modules
Genes in the selected modules were subjected to functional enrichment analysis with corresponding databases.The Gene Ontology database (GO,http://geneontology.org/) was used to identify genes and gene enrichment,and the Kyoto Encyclopedia of Genes and Genomes database (KEGG,http://www.genome.jp/kegg/) was used for pathway analysis.A threshold ofPvalue ≤ 0.05 was used to define significant GO categories and KEGG pathways,and the top ten records were extracted for each analysis.Cytoscape software (https://cytoscape.org/) was applied to further visualize the modules of interest,and the 50 genes with the greatest intramodular connectivity were identified as hub genes.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from nerve tissues and SCs using TRizol reagent from Sigma.The RNA was treated with RNasefree DNase I and reverse transcribed with a Prime-Script RT Kit (TaKaRa,Dalian,China) according to the manufacturer’s instructions.Quantitative real-time polymerase chain reaction was performed with the SYBR Premix Ex Taq (TaKaRa).Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize mRNA expression.The primers used are listed inAdditional Table 1.
Schwann cells primary culture and transfection
Primary SCs were isolated,maintained,and transfected as previously described (Yao et al.,2016).Sciatic nerves were harvested from seven 1-day-old Sprague-Dawley rats (specificpathogen-free) from the Experimental Animal Center of Nantong University and used to isolate SCs,which were then purified with anti-Thy1.1 antibody (mouse,1:1000,Cat# M7898,Sigma-Aldrich,St.Louis,MO,USA) and rabbit complement (25%,Cat# 31024100,Invitrogen,Carlsbad,CA,USA) to remove the fibroblasts.The purified SCs were maintained in Dulbecco’s modified Eagle medium (Gibco,Grand Island,NY,USA) containing 10% fetal bovine serum(Gibco) at 37°C under humidified conditions with 5% CO2.Primary cultured SCs were transfected with a negative control small interfering RNA (siRNA) or aKif22-specific siRNA(Ribobio,Guangzhou,China) using Lipofectamine RNAiMAX transfection reagent (Invitrogen),as previously described (Yao et al.,2016).The target sequences of the siRNA duplexes were as follows:Kif22-specific siRNA-1,5′-CCT GAA GAA AGG CCA CAA A-3′;Kif22-specific siRNA-2,5′-CCA TCA CTA GCT TCT CTG A-3′;Kif22-specific siRNA-3,5′-GTG ATT AAC CGG CCT TTC A-3′;negative control siRNA,5′-GGC UCU AGA AAA GCC UAU GC-3′.
5-Ethynyl-2′-deoxyuridine proliferation assay
SC proliferation was analyzed using a 5-ethynyl-2′-deoxyuridine(EdU) cell proliferation kit (Ribobio).The SCs transfected with the indicated siRNA were seeded into a 96-well culture plate at a density of 1 × 105cells/well and incubated with 50µM EdU for 4 hours.Next,the treated SCs were fixed in 4%paraformaldehyde for 30 minutes and permeabilized with Triton X-100 for 15 minutes.Then,the cells were stained with Apollo fluorescent dye,and the cell nuclei were stained with 5 µg/mL Hoechst 33342,for 20 minutes.The ratio of EdUpositive to total cells was calculated using ImageJ software(National Institutes of Health,Bethesda,MD,USA) from four randomly selected images photographed under a Olympus fluorescence microscope (Shinjuku,Tokyo,Japan).The EdU assay was carried out in triplicate,with three identical wells included for each sample.
Transwell assay
SC migration was detected using Transwell chambers (Corning,New York,NY,USA) with a diameter of 6.5 mm and with 8 µm pores,as previously reported (Yao et al.,2016).The bottom membrane of each chamber was coated with 5 µg/mL fibronectin (Millipore,Billerica,MA,USA) three times before use.Then,primary SCs resuspended in 100 µL Dulbecco’s modified Eagle medium (1 × 106cells/mL) were seeded to the upper chambers and 500 µL complete medium (Dulbecco’s modified Eagle medium with 10% fetal bovine serum) was added to the lower chambers.At the indicated time point,the cells were fixed with 4% paraformaldehyde for 30 minutes,and the cells that were adhered to the bottom chamber were stained with 0.1% crystal violet for 15 minutes.Next,the remaining crystal violet solution was remove by rinsing with sterile water,the cells were gently swabbed off of the upper chamber using a cotton swab.The stained cells were imaged under a Olympus fluorescence microscope.Finally,a 33%acetic acid solution was applied to dissolve the crystal violet,and a Synergy 2 multifunctional detector (BioTek,Burlington,VT,USA) was further used to detect the absorbance at 570 nm to quantify the migrated cells.Each experiment was performed in triplicate.
Wound healing assay
A wound healing assay was carried out using a Culture-Insert 2 Well in µ-Dish 35 mm (iBidi,Gräfelfing,Germany).The culture inserts were placed in 6-well plates before use.Then,SCs transfected with the indicated siRNA were seeded into the two wells of the culture inserts and maintained for 24 hours.Next,the culture inserts were removed to expose the blank area (or“wound”) between the two wells,which was photographed to serve as the control (0 hour) image.The remaining cells were starved in Dulbecco’s modified Eagle medium supplemented with 5% fetal bovine serum and 0.15 µg/mL mitomycin C(Sigma) for 12 hours.Finally,cells migrating across the wound in four randomly selected fields were photographed,and the percentage of wound healing (relative to the control image)was measured and analyzed using ImageJ software at the indicated time points.Three independent wells were included for each sample.
Western blot analysis
SCs transfected with the indicated siRNAs (Kif22-specific siRNAs or negative control siRNA) were lysed with a buffer containing protease and phosphatase inhibitors to extract total protein.The extracted proteins were subjected to electrophoresis on a 10% sodium dodecyl sulfatepolyacrylamide gel and then transferred to a nitrocellulose membrane (Merck Millipore,Billerica,MA,USA).Next,the membranes were incubated with anti-ERK (rabbit,1:1000,Cat# A4782,Abclonal,Boston,MA,USA),anti-p-ERK(phospho-ERK1-T202/Y204+ERK2-T185/Y187,rabbit,1:1000,Cat# AP0974,Abclonal),anti-protein kinase C α (PKCα;rabbit,1:1000,Cat# A11107,Abclonal),and anti-GAPDH (rabbit,1:2500,Cat# G9545,Sigma) antibodies at 4°C overnight.Then,the samples were incubated with goat anti-rabbit IgG-horseradish peroxidase (1:1000,Cat# abs20040,Absin Bioscience Inc,Shanghai,China) at room temperature for 2 hours.The blot images were scanned using a Tanon5200multisystem (Tanon,Shanghai,China).Finally,quantitative analysis was performed using ImageJ software.Protein levels were normalized to GAPDH.
Statistical analysis
Unpairedt-test was performed for all statistical analyses using GraphPad Prism 9 (GraphPad Software,San Diego,CA,USA).APvalue <0.05 was considered statistically significant,and all results are shown as mean ± standard deviation.
Results
Construction of co-expression modules for proximal and distal nerve segments from rats with sciatic nerve injury
To systematically analyze and compare gene expression between proximal and distal nerve segments,we first performed a microarray analysis and found that there were 17,482 and 13,988 differentially expressed genes at 0,1,and 6 hours,and 1,4,7,and 14 days post-sciatic nerve transection in the proximal and distal nerve stumps,respectively.We then used WGCNA,a method for transforming gene expression data into co-expression modules,to separately analyze the differentially expressed genes in the proximal and distal nerve stumps.Before constructing the co-expression modules,we filtered out differentially expressed genes with low expression variation (standard deviation ≤ 0.25),yielding 13,331 proximal and 10,726 distal genes.Power value is another critical parameter that affects the independence and average degree of connectivity of co-expression modules.Hence,we screened a range of power values and found that a power value equal to 14 resulted in high degrees of independence and average connectivity (Additional Figure 1).When this power value was used to generate the weighted co-expression network model,the 13,331 genes identified in the proximal segment were divided into 31 modules (Figure 1A).We next applied a Pearson correlation algorithm to calculate the correlation coefficient and P values of representative module genes and time to determine the importance of each proximal module(Figure 1B).In parallel,the 10,726 genes of distal segment were divided into 15 modules (Figure 2A),and the relative significance of the 15 modules was also determined based on Pearson correlation analysis (Figure 2B).The genes included in each module are listed inAdditional Table 2.The significant modules for both the proximal and distal segments are shown inFigures 1Band2B.
Functional enrichment analysis of genes in modules that correlate significantly with time
As described above,we used WGCNA to identify significant gene co-expression modules in proximal and distal nerve stumps from rats after sciatic nerve injury.Next,we performed KEGG and GO functional analyses of genes in selected modules to further understand the functional differences and similarities between the proximal and distal modules.There were significant differences in the KEGG pathways and biology processes that were enriched in the proximal and distal modules.The red proximal module was mainly enriched in genes related to axon guidance,extracellular matrix-receptor interaction,cell adhesion,and cyclic adenosine monophosphate signaling (Figure 3A),while the purple and blue proximal modules were mainly enriched in genes related to lysosomes,the cell cycle,DNA replication,and cytokine-cytokine interactions (Figure 3BandC).Most of these pathways have been previously reported to be involved in nerve regeneration.The distal modules were primarily enriched in genes involved in degeneration-related pathways such as calcium signaling,lysosomes,glycan degeneration,and fatty acid degeneration (Figure 3D–F).Consistent with this,GO enrichment analysis showed that the proximal modules were enriched in genes involved to nerve regeneration–related processes,and distal modules were enriched in genes related to nerve degeneration (Additional Figure 2).
To our surprise,genes related to inflammation,which is triggered by axonal degeneration distal to nerve damage and is an essential part of WD (Dubový et al.,2013),were enriched in the proximal modules instead of the distal modules that correlated positively with time (Figure 3CandAdditionalFigure 2A).To further investigate this apparent contradiction,we analyzed other modules.GO annotation analysis identified two modules that were associated with inflammation in distal segments (Additional Figure 3AandB).These two modules were positively correlated with time only 1 to 24 hours after injury,and were negatively correlated or uncorrelated with time until 14 days post-injury (Figure 2B).Many biological process categories,such as the inflammatory response,the immune response,wound healing,and cytokine-mediated signaling,were enriched in the two distal modules (Additional Figure 3AandB).These enriched categories are important for the onset of WD.These results are consistent with a previous study that suggested that there are three distinct phases within the 4-week post-injury period.The first two phases include 0 to 6 hours post-injury,which involves the acute response to injury,and 6 to 24 hours post-injury,which is associated with WD.The extended up-regulation of the inflammatory response observed in the blue proximal module may be associated with the induction of neuropathic pain following nerve injury.Some of the genes included in this module were previously reported to be related to neuropathic pain,such asCxcl13,Ccl2,andCxcr3.Thus,it appears that inflammation after nerve injury is important not only for promoting nerve regeneration,but also for improving functional recovery.
Next,we performed functional enrichment analysis of genes in the proximal and distal modules that were negatively correlated with time (Figure 4andAdditional Figure 4).As shown inFigure 4A–C,the three proximal modules were primarily enriched in genes involved in metabolism-related pathways such as fatty acid elongation,unsaturated fatty acid biosynthesis,and nitrogen metabolism.Axon regeneration after injury requires a large supply of energy,so these functions,as well as those of other metabolic pathways that store or consume large amounts of energy,may be down-regulated to help the damaged neurons successfully regenerate without undergoing an energy crisis.Two distal segment modules were also enriched in genes related to metabolism (Figure 4DandE).The relevant KEGG pathways are listed inAdditional Tables 3and4.Another distal module,the purple module,was enriched in genes involved in inflammatory-and apoptosis-related signaling pathways,which is consistent with the results from the GO enrichment analysis described above (Figure 4F).As discussed above,although genes in the purple distal module were negatively correlated with time overall,they were up-regulated at 1 and 6 hours post-injury,presumably due to participation in the initiation of WD.Furthermore,several biological processes were enriched in both proximal and distal modules (AdditionalFigure 5).In brief,processes associated with response to injury such as wound healing and drug response were upregulated in both proximal and distal segments (Additional Figure 5A),while processes associated with energystoring metabolic processes such as cholesterol and sterol biosynthesis were down-regulated in both segments over time(Additional Figure 5B).Taken together,our results suggest that peripheral nerve repair is a dynamic process that requires analysis at different time points,and that WGCNA can provide comprehensive insight into this process.
Identification of key genes related to axon regeneration and wallerian degeneration
To identify key genes regulating axon regeneration and WD,we selected the blue proximal module and the turquoise distal module for further analysis.In total,544 and 264 critical genes that were positively associated with time were identified in the blue proximal module and the turquoise distal module,respectively,based on an MM threshold of >0.9 and a GS threshold of >0.5 (Figure 5AandBandAdditionalTable 5).Next,Cytoscape software was used to calculate intramodular connectivity within each of the two modules and identify the 50 genes in each module with the greatest degree of intramodular connectivity,which we designated as hub genes (Figure 5CandD).Interestingly,we found thatNgfrandKif22were not only critical genes associated with time (Additional Table 5),but were also hub genes in both the blue proximal module and the turquoise distal module(Figure 5CandD).Ngfr,or nerve growth factor receptor,has been previously shown to be up-regulated in SCs after nerve injury,and to interact synergistically with nerve growth factor to guide axon regeneration and activate autophagy in SCs to enhance myelin debris clearance during WD,thus playing important role in peripheral nerve repair (Zhou and Li,2007;Webber et al.,2008;Jessen and Mirsky,2016;Li et al.,2020b).Kif22is a microtubule positive-end tracking protein that is crucial for cell mitosis,centrosome separation,and spindle formation (Yu et al.,2020).Previous studies have shown thatKif22promotes cancer cell proliferation and metastasis (Yu et al.,2014,2020).However,the role ofKif22in peripheral nerve repair is currently unknown.To verify the microarray and WGCNA results forKif22,we performed quantitative realtime polymerase chain reaction and found thatKif22mRNA expression was significantly up-regulated from 4 days postnerve injury in both the proximal and distal nerve stumps(Figures 1A,5Eand5F),indicating a potential role forKif22in axon regeneration and WD.Collectively,these results suggest thatKif22activity is an important part of peripheral nerve repair.
Kif22 regulates SC proliferation and migration via PKCαmediated ERK signaling
SCs,which are the most important glial cells in the peripheral nervous system,play an essential role in peripheral nerve development and function,and especially in peripheral nerve repair after injury.After injury,myelinating and nonmyelinating (Remak) SCs de-differentiate,activate,or reprogram to convert into repair SCs (Jessen and Mirsky,2016).Then,the repair SCs further proliferate,migrate,form bands of Büngner to guide axon regeneration,and secrete a variety of cytokines and neurotrophic factors to activate macrophages to engulf myelin fragments,thereby initiating WD and promoting nerve regeneration (Jessen and Mirsky,2016;Jessen and Arthur-Farraj,2019).Given the role of SCs in axon regeneration and WD and the critical role ofKif22in regulating cancer cell proliferation and metastasis,we next asked whether the effect ofKif22on SC proliferation and migration promotes peripheral nerve repair.First,we designed threeKif22-specific siRNAs to knock downKif22expression in SCs.As shown inFigure 5A,all threeKif22-specific siRNAs significantly reducedKif22expression.We used the two most effectiveKif22-specific siRNAs to silenceKif22in SCs and investigate the effect ofKif22knockdown on SC proliferation and migration.An EdU assay showed thatKif22knockdown decreased the rate of SC proliferation (Figure 6BandC),and a Transwell assay showed thatKif22knockdown disrupted SC migration (Figure 6DandE).The wound healing assay further validated the negative effect ofKif22knockdown on SC migration (Figure 6FandG).Taken together,these results suggest thatKif22promotes SC proliferation and migration.
We next explored the mechanisms underlyingKif22-mediated regulation of SC proliferation and migration.We first analyzed the effects ofKif22knockdown on the expression of genes involved in cell cycle and migration.In general,Kif22knockdown stably decreased the expression of several genes involved in the cell cycle,including cyclin dependent kinase 2 and cyclin D1 (Figure 7A).Cyclin D1 was identified as a critical gene associated with time in both the blue proximal module and the turquoise distal module based on an MM threshold of >0.9 and a GS threshold of >0.5 (Additional Table 5).Consistent with this finding,cyclin D1 has been previously reported to be down-regulated byKif22silencing,thereby inhibiting the proliferation of gastric cancer cells (Yu et al.,2020).These findings strongly suggest thatKif22knockdown can drastically decreased the expression of cyclin dependent kinase 2 and cyclin D1 to disrupt SC proliferation.Kif22knockdown also significantly reduced the expression of genes involved in cell migration,such as vimentin and cadherin 2(Figure 7B).These two genes encode key adhesion molecules that regulate SC migration (Clements et al.,2017;Yao et al.,2020).Therefore,our findings suggest thatKif22knockdown inhibits SC migration by modulating the expression of vimentin and cadherin 2.
Next,we explored the mechanism by whichKif22contributes to altering the expression of genes related to the cell cycle and migration.The ERK signaling pathway plays an important role in SC proliferation and migration,as well as regulation of axon regeneration and WD (Napoli et al.,2012;Li et al.,2020a).The ERK signaling pathway is regulated by numerous upstream signals,including epidermal growth factor,insulin-like growth factor,and other growth factor signals,as well as PKCα (Yin et al.,2013;Sun et al.,2015).A previous study found that PKCα is up-regulated in rat SCs from distal sciatic nerve stumps after injury and activates the ERK signaling pathway to promote SC proliferation and migration (Li et al.,2020a).However,here we found that PKCα was down-regulated byKif22knockdown in SCs (Figure 7C–E).Moreover,a previous study reported thatKif22is related to the ERK signaling pathway,and thatKif22knockdown reduces p-ERK expression levels in gastric cancer cells (Yu et al.,2020).Thus,we asked whether alteringKif22expression would affect activation of the ERK signaling pathway in SCs.Western blot analysis showed that knocking downKif22in SCs withKif22-specific siRNAs significantly inhibited ERK phosphorylation (Figure 7DandE).Taken together,these date indicate thatKif22modulates SC proliferation and migration by affecting PKCα expression and thereby activating the ERK signaling pathway.
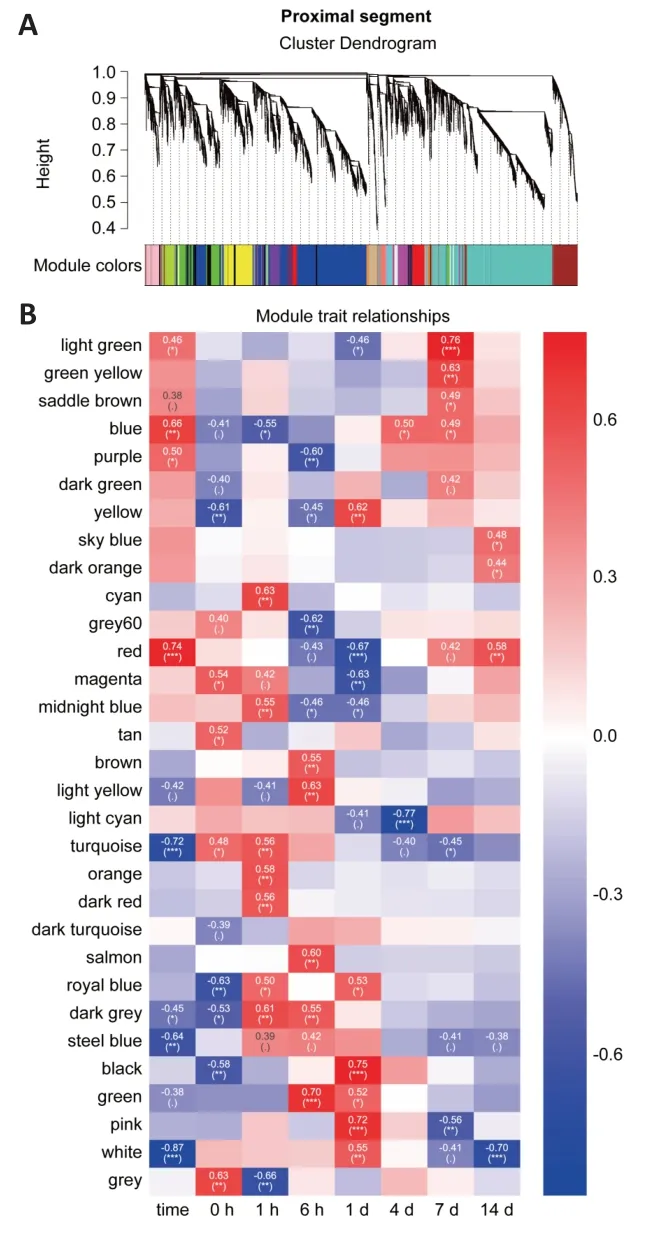
Figure 1|WGCNA and ide ntification of significant DEG modules from proximal nerve segments.

Figure 2|WGCNA and identification of significant DEG modules from distal nerve segments.

Figure 3|Functional KEGG analysis of gene enrichment in modules that correlate positively with time.

Figure 4|Functional KEGG analysis of genes enriched in modules that correlate negatively with time.
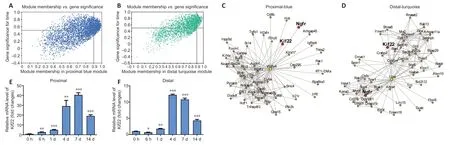
Figure 5|Kinesin family member 22 (Kif22) is the key hub gene that correlates positively with time in both the proximal and distal segments.

Figure 6|The effect of Kif22 knockdown on SC proliferation and migration.
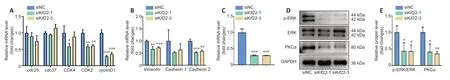
Figure 7|Kif22 knockdown decreases PKCα expression,resulting in inactivation of the ERK signaling pathway.
Discussion
Peripheral nerve injury repair is a complex process that is dependent on effective WD and,subsequently,successful axon regeneration.In this study,we applied WGCNA to systematically analyze the transcriptomes of proximal and distal nerve stumps from rats subjected to sciatic nerve injury,with the aim of better understanding the cellular and molecular mechanisms that underlie the coordinated regulation of axon regeneration and WD in peripheral nerve injury repair.
Using WGCNA,we identified 31 and 15 co-expression modules in proximal and distal nerve segments,respectively,from rats with sciatic nerve injury.We then selected the most significant co-expression modules and performed KEGG and GO enrichment analyses of the genes in these modules to further understand the functional differences and similarities between the two segments.The results from these analyses showed that nerve regeneration-related biological processes were up-regulated in proximal modules,whereas nerve degeneration-related biological processes were up-regulated in distal modules.Genes associated with inflammation were enriched in both the proximal and distal modules.However,the inflammation-related genes in the proximal modules were positively correlated with time,while those in the distal modules were negatively correlated with time,and were first up-regulated and then down-regulated.This may reflect an important difference between the mechanisms involved in axon regeneration and WD.In addition,the proximal and distal modules showed some similarities in terms of enriched biological processes,including up-regulation of genes involved in processes associated with response to injury such as wound healing and drug response,as well as downed-regulated processes associated with energy-storing metabolism such as cholesterol and sterol biosynthesis.Taken together,these findings suggest ways in which regeneration and degeneration could be targeted separately or together for therapeutic purposes.
In this study,we identifiedKif22as a hub gene that is positively associated with time in both proximal and distal segments.Kif22is a member of the kinesin-like family of proteins,which are microtubule-dependent molecular motors with DNA-binding capacity (Yu et al.,2014).Kif22has been previously reported to promote cancer cell proliferation and migration (Yu et al.,2014,2020;Pike et al.,2018).In addition,we found thatKif22knockdown substantially inhibits SC proliferation and migration,indicating a potential role forKif22in axon regeneration and WD.Furthermore,Kif22knockdown reduced the expression of cell cycle-and migration-related genes,as well as the expression of PKCα.PKCα is a positive regulator of the ERK signaling pathway,which promotes SC proliferation and migration (Li et al.,2020a).Consistent with this,reduced expression of PKCα inKif22-knockdown SCs led to delayed activation of the ERK signaling pathway.ThoughKif22has been previously reported to affect ERK activation,the mechanism underlyingKif22-mediated regulation of the ERK signaling pathway has not yet been determined (Yu et al.,2020).The findings from our study suggest thatKif22regulates the ERK signaling pathway by modulating PKCα expression.However,the molecular mechanism by whichKif22regulates PKCα expression remains unclear.Kif22was previously shown to not only act as a motor protein during cell mitosis,but also transcriptionally regulate cancer development (Yu et al.,2014).Thus,we speculate thatKif22regulates PKCα transcription,which should be explored in future studies.Collectively,these data suggest a new role forKif22in peripheral nerve injury repair and a new mechanism by whichKif22regulates the ERK signaling pathway.
The current study suggests thatKif22mechanistically regulates nerve repair by modulating ERK signaling-mediated SC proliferation and migration.However,as an important regulator of cell division,proliferation,and migration,Kif22may not only act on nonnneuronal cell proliferation but also be repurposed from its mitotic role to play a cytoplasmic role in neurons.Kif22is a motor protein that can regulate microtubule dynamics to delay epidermal growth factor receptor internalization and enhance epidermal growth factor receptor signaling (Pike et al.,2018),and the findings from our study suggest that it may also regulate the transport of growth factors to regulate nerve regeneration.Microtubules,as an important part of the cytoskeleton,play essential roles in nerve regeneration (He et al.,2016;Blanquie and Bradke,2018);thus,Kif22may regulate microtubule dynamics to participate in nerve repair.Future studies should explore these possible molecular mechanisms underlyingKif22-modulated nerve repair.
In summary,this study provides a comprehensive comparison of proximal axon regeneration and distal axon WD.We found thatKif22is a positive regulator of SC proliferation and migration,processes that are essential in axon regeneration and WD,highlighting potential new therapeutic targets for treating peripheral nerve injury.However,while we verified thatKif22mechanistically regulates nerve repair by modulating ERK signaling-mediated proliferation and migration of SCs,it remains unclear whether the role ofKif22in mitosis,microtubule dynamics,or transport of growth factors is also a part of this process.In addition,we only focused on hub genes with the same expression pattern in both proximal and distal segments,so hub genes with different expression patterns in these segments merit further study.
Author contributions:Study conceptualization:QQC‚BY;study design‚data analysis and manuscript writing:QQC;experiment implementation:YL‚QS;technical assistance:ML‚XAN;English writing assistance and manuscript review:SSM.All authors read and approved the final version of the manuscript.
Conflicts of interest:The authors declare no conflict of interest.
Financial support:This study was supported by the National Major Project of Research and Development of China‚No.2017YFA0104701 (to BY);the National Natural Science Foundation of China‚No.32000725 (to QQC);the Natural Science Foundation of Jiangsu Province of China‚No.BK20200973 (to QQC);and the Jiangsu Provincial University Innovation Training Key Project of China‚No.202010304021Z (to ML).The funders had no role in study design‚data collection and analysis‚decision to publish‚or preparation of the manuscript.
Institutional review board statement:This study was approved by the Institutional Animal Care guidelines of Nantong University‚China(approval No.S20210322-008) on March 22‚2021.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal‚and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License‚which allows others to remix‚tweak‚and build upon the work non-commercially‚as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Table 1:List of primers used in quantitative real-time polymerase chain reaction.
Additional Table 2:The number of genes in different modules of proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration.
Additional Table 3:The list of enriched KEGG pathway related to Figure 3.
Additional Table 4:The list of enriched KEGG pathway related to Figure 4.
Additional Table 5:List of moddule membership and gene significance in proximal blue and distal turquoise modules.
Additional Figure 1:Analysis of network topology for various soft-thresholding powers.
Additional Figure 2:Functional GO enrichment of genes in modules that positively correlated with time.
Additional Figure 3:Functional enrichment of genes in modules that first positively .then negatively correlated with time.
Additional Figure 4:Functional enrichment of genes in modules that negatively correlated with time.
Additional Figure 5:The overlap between the biological function of proximal and distal segments.
杂志排行
中国神经再生研究(英文版)的其它文章
- Long non-coding RNA MEG3 regulates autophagy after cerebral ischemia/reperfusion injury
- Social skills and psychopathology are associated with autonomic function in children:a cross-sectional observational study
- The mechanism behind activation of the Nod-like receptor family protein 3 inflammasome in Parkinson’s disease
- Effects of electroacupuncture on pain sensation in a rat model of hyperalgesia with nicotine dependence
- HOTTIP downregulation reduces neuronal damage and microglial activation in Parkinson’s disease cell and mouse models
- Involvement of A5/A7 noradrenergic neurons and B2 serotonergic neurons in nociceptive processing:a fiber photometry study