Comparison of DNA Extraction Methods for Four Kinds of Seafood in Qinhuangdao
2022-07-13ShanshanLIUHaijuanZHANGWeiweiWANGDongmeiZHAOZhichengWANGYingLIU
Shanshan LIU Haijuan ZHANG Weiwei WANG Dongmei ZHAO Zhicheng WANG Ying LIU




Abstract[Objectives] This study was conducted for the purpose of widely using molecular biology techniques in the detection of seafood species. [Methods]In this study, such four kinds of seafood as fish, shrimps, crabs and shellfish were used as samples to compare three DNA extraction methods, including Tiangen marine animal tissue genome extraction kit method (DP324)(a), SN/T 3589.6-2013/8.3(b) method and modified SN/T 3730.8-2013/8.1(c) method. The purity and concentrations of DNA and the amplification effects of real-time fluorescene PCR were compared. [Results] The three methods had different extraction effects while satisfying the real-time fluorescent PCR detection. The DNA templates extracted by the SN/T 3730.8-2013/8.1 standard method had higher purity and concentrations, and showed clearer bands in gel electrophoresis, indicating that the method has less effect on real-time fluorescent PCR and less inhibition, and can meet the detection needs of the four types of seafood, including fish, shrimps, crabs and shellfish. [Conclusions]This study provides a theoretical basis for the detection of seafood animal-derived components.
Key wordsSeafood; DNA extraction; Real-time fluorescence PCR; Detection
Qinhuangdao City is an important tourist city in China's Bohai Rim region. It is a region with a wide variety of seafood such as shellfish, fish, crabs, and shrimps. Because some seafood varieties are similar in shape or difficult to accurately identify after processing, misjudgment can be caused easily, thereby causing economic losses to consumers and producers. The molecular biology technology has high sensitivity and specificity and has been widely used in the identification of biological species[1-5], while the quality of DNA extracted in the early stage is an effective guarantee for subsequent experiments, so a fast, efficient, economical and safe DNA extraction method suitable for different samples is very important for subsequent experiments such as polymerase chain reaction (PCR), real-time fluorescence PCR, enzyme digestion and species identification.
Smith[6] compared a variety of DNA extraction methods from potatoes and their products. The CTAB method was used to extract DNA from tuber materials with the highest yield, and the Wizard method was suitable for DNA extraction of most potato processed products, but no universally applicable method for DNA extraction was found. Studies have found that the residues of extraction reagents[7], temperature[8], physical effects such as grinding and shearing[9], different samples and different processing methods of the same sample[10-12] will both affect the quality of DNA extraction, and then affect subsequent identification tests[13].
Preliminary experiments found that, different from the above-mentioned animal organs, bacteria, and blood, seafood samples are rich in amino acids, fatty acids, proteins and other macromolecular substances, and the extraction of DNA is easily affected by freshness and extraction process. It is of great significance to find a DNA extraction method that is suitable for a variety of seafood and satisfies real-time fluorescent PCR detection for the purpose of detecting the source components of various types of seafood and their products in large quantities[14-16].
In this study, three methods were compared for the effect of DNA extraction, namely Tiangen marine animal tissue genome extraction kit method (DP324) (method a), SN/T 3589.6-2013/8.3 Identification of fish species in export food―Part 6: Detection of tuna fish ingredient―Real-time PCR method (method b) and the modified method of SN/T 3730.8-2013/8.1 Identification of domestic animal ingredient in food and feed―Part 5: Detection of pig ingredient―Real-time PCR method (method c). Since previous studies found that the SN/T 3730.8-2013/8.1 method has a long centrifugation time and is not suitable for the extraction of seafood samples, it has been improved, and the four-step centrifugation of "8.1.3, 8.1.4, 8.1.7, 8.1.8" in the method was adjusted to 10, 5, 5 and 5 min, which was called "Modified SN/T 3730.8-2013/8.1", that is, method c. The concentrations and purity of DNA extracted by the three methods were determined, and the significance of differences between species and methods were analyzed by the SPSS 22.0 software. The DNA extracted was analyzed by agarose gel electrophoresis for quality, and subjected to real-time fluorescence PCR reaction with 18S rRNA primer probe[17], and the amplification effects were compared to find a DNA extraction method suitable for seafood samples and provide guarantee for the detection of seafood animal-derived components.
Materials and Methods
Experimental materials
Sample preparationFresh fish, shrimps, crabs and shellfish samples were purchased from Qinhuangdao City and manually processed. The sampling sites are shown in Table 1. The tissues were placed in 2 ml centrifuge tubes and frozen at -80 ℃ for later use.
ReagentsCTAB, EDTA and Tris were purchased from Beijing Coolaber Technology Co., Ltd. Marine animal tissue genomic DNA extraction kit, TE buffer, proteinase K and RNA enzyme were purchased from Tiangen Biotech (Beijing) Co., Ltd. Other chemical reagents were purchased from Tian Jin Hui Hang Hua Gong Technology Co., Ltd. All reagents used in this method were of analytical grade.
Instruments and equipmentFresco21 desktop refrigerated high-speed centrifuge, Thermo Fisher Scientific; BS224S electronic balance, Beijing Sartorius Instrument Systems Co., Ltd.; CK1000D high-throughput tissue grinder, Beijing Tomorgen Biotechnology Co., Ltd.; MX-S vortex mixer, SCILOGEX, USA; NanoDrop micro-spectrophotometer, Thermo Fisher Scientific; S1010 mini centrifuge, SCILOGEX, USA; DTH-100 dry type thermostat, Hangzhou MIULAB Instrument Co., Ltd.; real-time fluorescence PCR detection system: AB Applied Biosystems, Step One Real-Time PCR System; gel imaging system: aplegen, omega fluor plus.
Experimental methods
DNA extractionThe three DNA extraction methods are shown in Table 2.
Evaluation of DNA extraction effectThe concentration and purity of the extracted DNA from the four types of marine samples were determined and subjected to 1% agarose gel electrophoresis, and the imaging observation was performed using a gel imager.
Real-time fluorescence PCR
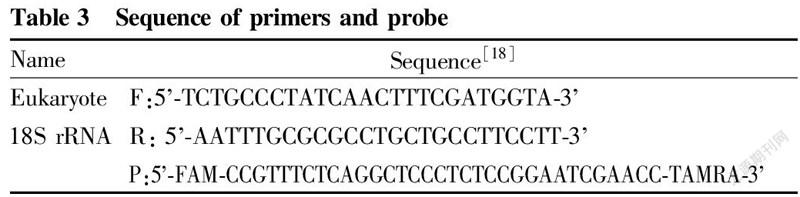
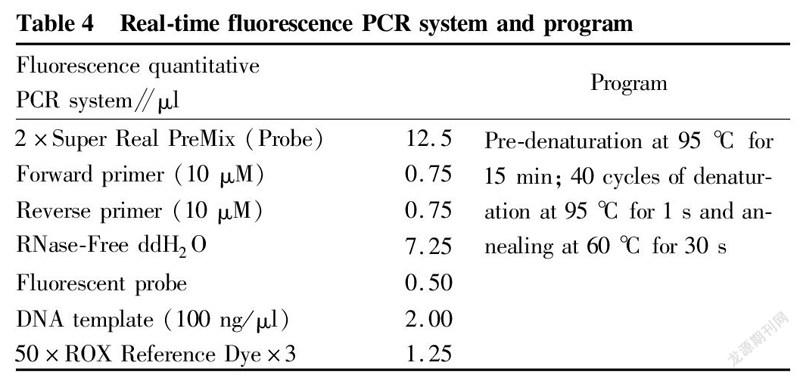
The extracted DNA was detected by real-time fluorescence quantitative PCR. The primer probe sequences used are shown in Table 3, and the real-time fluorescence PCR reaction was carried out according to the system and program described in Table 4.
Data analysis
The SPSS 22.0 statistical software was used to perform one-way analysis on the DNA purity of different extraction methods for each species above, as well as the DNA purity of different species in the same method, and the DNA concentrations of different species in different extraction methods were compared at the same time. Since there was no correlation between the four types of samples and they were relatively independent samples, the Kruskal-Wallis K method was used for significance analysis, and the P value was obtained. When the P value was less than 0.05, the difference was significant; when the P value was less than 0.01, the difference was extremely significant; and when P was greater than 0.05, it meant that there was no significant difference.
Results and Analysis
DNA purity
The purity of DNA extracted by the three methods (A260/A280) measured by NanoDrop microspectrophotometer is shown in Table 5. The ratio between 1.7 and 1.9 indicated higher purity, while a lower ratio might be due to the presence of macromolecular substances (polyphenols, protein, etc.) contamination, and a higher ratio might be due to the degradation of nucleic acids[19]. The A260/A280 ratio of method a was about 1.75 ± 0.05, and the purity was within the normal range; the purity ratio of method b for extracting DNA was higher, which might be because that the method caused nucleic acid degradation during extraction; and the A260/A280 of method c was 1.77 ± 0.05, so the purity was higher. The results of difference analysis showed that the purity of the four types of seafood DNA was significantly different when extracted by the three methods. Among them, the differences of the eight species were the most significant when using method a, followed by method b, and the differences of method c were the smallest. When the same sample was extracted by different methods, the purity of shrimp and crabs was significantly different, but there were no significant differences in fish and shellfish. The reason might be that the chemical compositions of the sampling parts of fish and shellfish were relatively consistent, and the extraction was not easily affected by lysing solutions, extracting solutions and mechanical force in different methods, so there was little difference between the methods. However, the muscle tissues of shrimps are easy to form gel[20-21] when they are crushed, which is not conducive to lysis and extraction, so method c with two steps of precipitation was effective. Other two methods showed different species exceeding the appropriate concentration range, resulting in significant differences between methods. For crab legs with higher water content, the extraction effect of method a was better. Meanwhile, it was found that method a and method c both showed two species with concentrations out of the suitable range, and there were many species in method b that were not within the suitable range, indicating that the applicability of method b was lower than that of other two methods. Based on the above analysis, it is better to use method a and method c to obtain the genomic DNA of multiple species and large-volume samples that meets the requirements of concentration.
DNA concentration
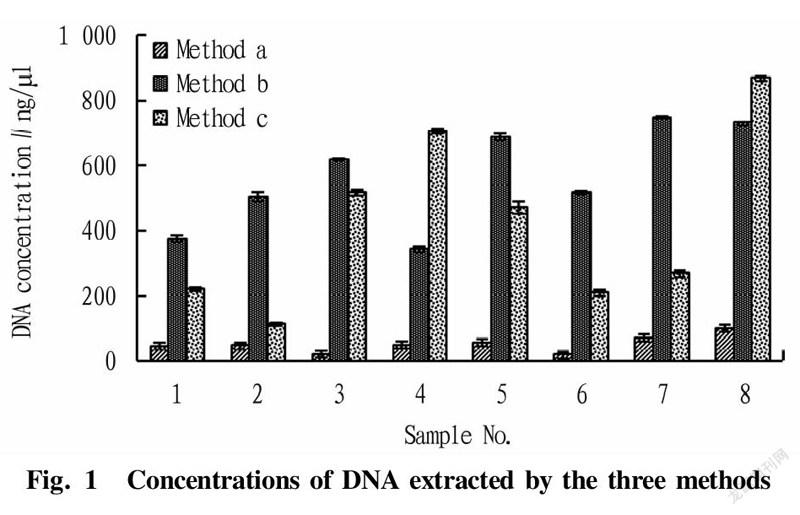
Comparison on the concentration results of the DNA extracted by the three methods is shown in Fig.1. It can be seen from Fig. 1 that the concentrations of DNA extracted by method b and method c were higher, and the scallop sample extracted by method c had the highest concentration. The concentrations of DNA extracted by method a were generally low for different species. The reason might be that method a is a centrifuge column method, in which after DNA is bound with the centrifuge column, there will inevitably be residues even after elution, and even in some species, the losses are relatively large. Meanwhile, it was found that the lysis speed of method a was faster, but the extraction concentrations were not high, which might be due to that the lysis solutions and the polyphenols and amino acids in the seafood samples[22] affected the binding of DNA to the centrifuge column, resulting in lower final concentration results. Method b and method c had their own advantages in the extraction effects of different species, and the extraction concentrations of shellfish skirt were generally higher. Specifically, the DNA concentrations of scallop skirt obtained by method c were the highest, and the extraction results of DNA from yellow clams by method b were the second. Therefore, with DNA concentration as an index, and the extraction effects of method b and method c were better.
The extracted DNA was subjected to agarose gel electrophoresis and imaged under ultraviolet light. The results are shown in Fig. 2.
As shown in Fig. 2, there were significant differences in the clarity and brightness of DNA bands extracted by the three methods. Method a showed poor clarity of bands, and individual samples had no bands. The samples of method b had clearer bands but more short fragments at the front of electrophoretic bands, indicating that the DNA was degraded. The bands of method c were the clearest and brightest, because method c had two times of isopropanol precipitation, the impurities in templates were less, and the centrifugation time of the improved method was greatly shortened, which avoided the degradation of DNA.
Real-time fluorescence PCR detection
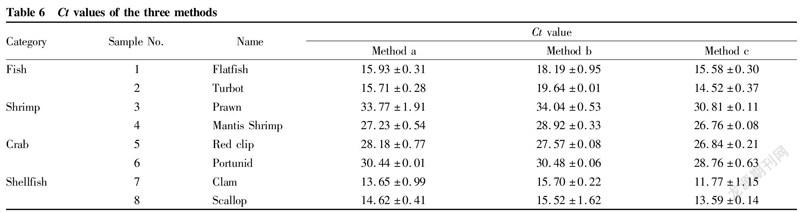
In order to compare the efficiency of real-time fluorescent PCR between different methods, this study used eukaryotic universal 18S rRNA as the target gene for determination, and the Ct values obtained are shown in Table 6.
The results in Table 6 showed that the Ct values of method c were generally lower for different species. Except for two crab samples with Ct values greater than those of method b, remaining Ct values of method a were between method b and method c. The Ct values of method b were higher. The values of individual samples were larger than 30, and that of one sample was even larger than 35, which might be misjudged as negative. From the above analysis, it could be concluded that although the extraction concentrations of individual samples in method c were not the highest, due to the high DNA quality after two times of isopropanol precipitation, the amplification fluorescence signal was stronger, and the final detection Ct values were low, which avoided false negative results.
Comparison of the three extraction methods
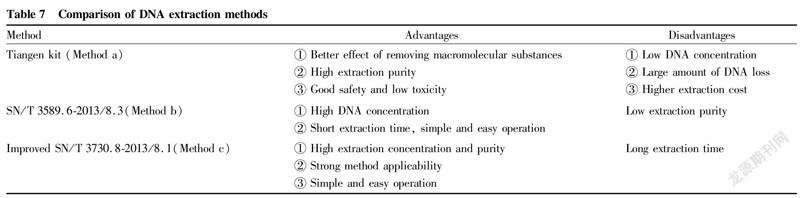
Through data analysis and identification experiments, the advantages and disadvantages of the three extraction methods are summarized in Table 7.
Conclusions and Discussion
It is necessary to select an appropriate DNA extraction method based on the analysis of the differences of DNA purity and different concentrations. High-quality DNA template is a key prerequisite for PCR amplification, product analysis and sequencing, species identification, etc., and is jointly determined by two factors, DNA concentration and purity. Residues of extractants such as ethanol directly affect PCR amplification, so DNA concentrations must be within an appropriate range to meet the requirements of subsequent experiments. In terms of DNA purity, different experiments have different requirements. Generally, higher DNA concentrations are more suitable for PCR amplification and subsequent species identification experiments. However, the concentration of templates required by fluorescence quantitative PCR is generally required to be between 10 and 100 ng/μl. Therefore, instead of simply relying on the concentration of DNA as the most important factor for evaluating the extraction effect, it is necessary to comprehensively select extraction methods according to the types of experiments combined with the purity of DNA.
Comparing the electropherograms with the DNA concentration extraction results, it was found that although the DNA concentrations extracted by method b were higher, they did not match the brightness of the electrophoretic bands, and more small degraded fragments appeared. These small fragments could not increase the true concentration of DNA and could easily lead to the failure of subsequent PCR amplification. Therefore, the concentrations of DNA templates measured by instruments alone are not reliable data, and electrophoresis verification should be carried out if subsequent experiments are required.
The DNA extraction of seafood samples were difficult to extract due to the effects from the polypeptides, polysaccharides, unsaturated fatty acids and other substances of the species themselves, and meanwhile, the extracted DNA templates were easily degraded, which made subsequent PCR tests prone to false negatives. Previous studies have focused on DNA extraction from a certain species or different products of the same species, and there are also studies on the extraction effects of certain methods on different species, but few reports are about the comparison of multiple methods for multiple samples. This study detected derived components of various marine samples, and compared the applicability of the three methods, aiming to find a simple and easy method and provide methodological guidance for the subsequent detection of derived components in multiple species in large quantities. Through DNA extraction from eight common species of fish, shrimps, crabs and shellfish in Qinhuangdao sea area, three DNA extraction methods were compared with DNA concentration, purity and real-time PCR efficiency as indices. The results showed that the extraction purity of the Tiangen kit method and the modified SN/T 3730.8-2013/8.1 method was higher, and the extraction concentrations of the SN/T 3589.6-2013/8.3 and the modified SN/T 3730.8-2013/8.1 methods were higher. In the real-time fluorescent PCR detection, the amplification curve signal of the modified SN/T 3730.8-2013/8.1 method was the strongest, and the Ct values were the smallest. In summary, under the premise of preventing contamination, the modified SN/T 3730.8-2013/8.1 method is more suitable for DNA extraction of four kinds of seafood. This method is simple and convenient to operate, and can obtain DNA with high concentrations and purity. It is suitable for real-time fluorescence PCR detection with good amplification efficiency, and provides a guarantee for subsequent identification tests.
In the study, it was found that the freshness of seafood samples also has a great influence on the quality of DNA extraction. If we want to obtain template DNA with higher concentrations and purity, we should try to choose fresher samples for extraction. Meanwhile, the final extraction results were also different due to different sampling parts of the samples. The concentrations extracted from the gills of fish, shrimp and crab samples were generally higher than those of other parts, and the concentrations extracted from the skirt of crabs were higher. However, since the ultimate purpose of this study is to apply the methods to the detection of food species-derived components, during which the gills and other parts are generally been removed from food samples, the parts that are easier to obtain in actual commercial samples were selected for research. In response to the above findings, the effects of the freshness of seafood samples and the sampling sites on DNA extraction remain to be further studied.
References
[1] WANG Y, JUNG JA, KIM WH, et al. Morphological and rDNA fluorescence in situ hybridizationanalyses of chrysanthemum cultivars from Korea[J]. Horticulture, Environment and Biotechnology, 2021, 62(6): 1-9.
[2] CHEN H, ZHAO XR, LIN QM, et al. Using a combination of PLFA and DNA-based sequencinganalyses to detect shifts in the soil microbial community composition after a simulated spring precipitation in asemi-arid grassland in China[J]. Science of the Total Environment, 2019, 657(C): 1237-1245.
[3] KAKAVAS VK. Sensitivity and applications of the PCR single-strand conformation polymorphism method[J]. Molecular Biology Reports, 2021, 48(4): 1-7.
[4] DAPRA V, ALLIAUDI C, GALLIANO I, et al. TaqMan real time PCR for the detection of the Gilbert's Syndrome Markers UGT1A1*28; UGT1A1*36 and UGT1A1*37[J]. Molecular Biology Reports, 2021, 48(5): 1-7.
[5] WANG JY, MI TZ, YU ZG, et al. Species-specific detection and quantification of scyphomedusae in Jiaozhou Bay, China, using a quantitative real-time PCR assay[J].Journal of Oceanology and Limnology, 2021, 39(4): 1360-1372.
[6] SMITH DS, MAXWELL PW, DE BSH. Comparison of several methods for the extraction of DNA from potatoes and potato-derived products[J]. Journal of agricultural and food chemistry, 2005, 53(26): 9848-9859.
[7] SHEN CF, ZHOU LH, PENG DW, et al. A rapid extraction method of tea genome DNA suitable for PCR amplification[J]. Journal of Tea Communication, 2020, 47(4): 593-596. (in Chinese).
[8] HU QL, JIANG Q, HUANG YL, et al. Optimization of DNA extraction methods for two species of Crabaea[J]. Molecular Plant Breeding, 2019, 17(2): 494-501. (in Chinese).
[9] SUN MM, HUANG HH, SHEN XF, et al. Comparison of different DNA extraction methods in soybean transgenic detection [J]. Jiangsu Agricultural Science, 2019, 47(4): 47-50. (in Chinese).
[10] YANG JR, XIONG X, HUANG MH, et al. Comparison of DNA extraction methods and species identification of fish meat[J].Modern Food Science and Technology, 2019, 35(6): 161-170. (in Chinese).
[11] HAN D, JIA XS, AN R, et al. The influence of processing on the DNA integrity in several raw materials of marine foods[J]. Periodical of Ocean University of China, 2013, 43(6): 58-63. (in Chinese).
[12] CHENG YH, PU Q, LU LX, et al. Effects of different food processing methods on DNA quality of Lophius lituion[J]. Journal of Food Safety and Quality, 2016, 7(6): 2509-2515. (in Chinese).
[13] ZHANG FM, LIAN SS, LIU SN, et al. Extraction of DNA from shell of bivalve mollusk and evaluation of its methodology[J]. Journal of Ocean University of China: Natural Sciences, 2020(S01): 7. (in Chinese).
[14] HUANG MH, XIONG X, XU WJ, et al. Development and validation of a duplex PCR for the simultaneous detection of Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) in processed fish products[J]. Modern Food Science and Technology, 2020, 36(12): 274-282. (in Chinese).
[15] XU WJ, XIONG X, HUANG MH, et al. Comparison of extraction effect of several DNA extraction methods in tuna products[J]. Modern Food Science and Technology, 2021, 37(7): 57-65. (in Chinese).
[16] XIONG X, GUARDONE L, GIUSTI A, et al. DNA barcoding reveals chaotic labeling and misrepr esentation of cod (Xue) products sold on the Chinese market[J]. Food Control, 2016(60): 519-532.
[17] General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China. Identification of domestic animal ingredient in food and feed―Part 5: Detection of pig ingredient―Real-time PCR method: SN/T 3730.8-2013[S]. Beijing: Standards Press of China, 2013. (in Chinese).
[18] STILWELL JM, ROSSER TG, WOODYARD ET, et al. Characterisation of myxozoan faunaof western mosquitofish, Gambusia affinis (Baird and Gerard) (Cyprinodontiformes: Poeciliidae), inhabiting experimental catfish ponds in Mississippi, USA[J]. Systematic parasitology, 2021, 98(4): 423-441.
[19] HAO YF, WANG HJ. Research progress of targeted protein degtadation technology[J]. Chemistry & Bioengineering, 2020, 37(6): 1-5. (in Chinese).
[20] LAN WB, MAO WJ, CHI AY. Study on protein composition and molecular weight distribution in muscle of Litopenaeus vannamei[J].Science and technology of food industry, 2012, 33(9): 49-52.
[21] YUAN LL, LIU SC, XIE WC, et al. Study on gel properties of shrimp meat and fish meat mixture [J]. Science and Technology of food industry, 2013, 34(12): 246-250. (in Chinese).
[22] TAGLIAVIA M, NICOSIA A, SALAMONE M, et al. Development of a fast DNA extraction methodfor sea food and marine species identification[J]. Food Chemistry, 2016(203): 375-378.
杂志排行
农业生物技术(英文版)的其它文章
- Report on the Breeding of Dahen 799 Broilers
- Evaluation and Analysis for Survey of Tea Production Quality Safety Management and Control
- Study on the Preparation of "Oil-tea" Instant Tea from the Compound Extract of Green Tea and Ginger
- Research Progress on Chemical Constituents and Pharmacological Effects of Zhuang Medicine Cocculus laurifolius DC.
- Study on Quality Standard of Lujing Yiqi Shengxue Pills
- Research on the Development of Guangxi Zhuang and Yao Ethnic Medicine Industry
