CRISPR/Cas Based Biosensing Platforms for Molecular Diagnosis
2022-04-22LITianLINa
LI Tian,LI Na
(1.School of Basic Medical Sciences,Henan University of Science and Technology,Luoyang 471023,China;2.College of Chemistry and Molecular Engineering,Peking University,Beijing 100871,China)
Abstract:Clustered regularly interspaced short palindromic repeats(CRISPR)and CRISPR-associated proteins(Cas)have become highly promising candidates in the construction of biosensing systems and diagnostic devices,in addition to their roles as revolutionary genome engineering tools.For in vitro diagnostics,the classⅡCRISPR/Cas systems possess unique advantages,such as high detection sensitivity and specificity,and the feasibility for developing point-of-care diagnostic technology.This minireview briefly introduces the working mechanism of CRISPR/Cas systems and mainly focus on the various diagnostic methods based on the CRISPR/Cas for detecting different forms of the genetic molecules in recent three years according to different readout methods.
Key words:CRISPR/Cas;molecular diagnosis;point-of-care testing;biosensing
Molecular diagnosis is one of the important frontier fields in the development of contemporary medicine,and its core technology is gene diagnosis.Acting as the building block of all organisms’genetic information,nucleic acids carry distinctive and unique feature of many diseases,including infection,cancer,autoimmune and genetic diseases.Rapid,accurate and cost-effective detection of the existence or mutation of nucleic acids is of great significance for disease prevention,prediction,diagnosis,treatment and prognosis.Due to the high sensitivity and specificity,polymerase chain reaction(PCR)is considered as the gold standard method in laboratories and clinical diagnosis for nucleic acid-based detection.It is surely good to carry out accurate testing in hospitals or professional testing institutions,which have well-equipped laboratory and skilled medical personnel.However,the outbreak of Corona Virus Disease 2019(COVID-19)make us more deeply realize that it is essential to develop point-of-care(POC)tests because the healthcare system will experience serious challenge during a pandemic.A POC test,which meet the“ASSURED”criteria(affordable,sensitive,specific,user-friendly,rapid and robust,equipment-free,deliverable to end users)set by the World Health Organization and can be conducted by any user at any given time and place in the absence of professional instruments,is vital for effective disease treatment and management[1].
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein(CRISPR/Cas)is an adaptive immunity system originally derived from most bacteria and archaea to defend the invasion of nucleic acids.The gene editing technology based on CRISPR/Cas system is hailed as the biggest biological science breakthrough in this century with its editability and high accuracy,which has had various applications ranging from fundamental research to clinical medicine,including generation of animal models,functional gene screening,epigenetic control,cellular genome imaging and treatment for genetic disease.In addition to their roles as revolutionary genome engineering tools,CRISPR/Cas systems are also highly promising candidates in the construction of biosensing systems and diagnostic devices,which have attracted significant attention recently.Although the majority of the biosensing platforms based on CRISPR/Cas systems are developed for nucleic acids,this technology can also be applied to non-nucleic acid target detection in combination with functional nucleic acids and molecular translators[2-6].
In recent years,CRISPR/Cas have been extensively investigated and several CRISPR/Cas systems have been developed as powerful gene editing tools.The fundamentals of CRISPR/Cas systems and their applications have been summarized in quite a few elegant articles[7-11].CRISPR/Cas systems based biosensing platforms have also been reviewed from different scientific and technical perspectives[1,6,11-14].In this minireview,we briefly introduce the working mechanism of CRISPR/Cas systems and mainly focus on the various diagnostic methods based on the CRISPR/Cas system for detecting different form of the genetic molecules in recent three years according to different readout methods.
1 Working mechanism of common CRISPR/Cas systems
CRISPR/Cas is an immune mechanism derived from bacteria and archaea, which is firstly discovered in 1987. The bacteria constantly collect the genetic information of viruses and integrate them into CRISPR.Once the same virus invades,they can recognize the exogenous DNA and cut them off. With the development of biological information technology,more and more CRISPR systems in bacteria have been excavated.The CRISPR/Cas systems are divided into two classes according to the composition of effector modules.In classⅠ,the effector module consists of several proteins with different functions.While in classⅡ,the effector module is only related to a single protein(Fig.1).

Fig.1 Fundamental components of CRISPR/Cas9,CRISPR/Cas12a,CRISPR/Cas14,and CRISPR/Cas13a systems[15]pink triangles indicate cis-cleavage sites
The ClassⅡCRISPR system is widely used and the most famous subtype is CRISPR/Cas9(typeⅡ).In a typical CRISPR/Cas9 system,the Cas9 nuclease efficiently shears the exogenous double-stranded DNA(dsDNA)sequence upon recognition of specific complementary dsDNA containing the 5′-NGG-3′protospacer adjacent motif(PAM)sequences in the presence of a guiding RNA(gRNA).ClassⅡCRISPR/Cas systems also encompass other types,such as type V,including Cas12 and Cas14,and typeⅥ,including Cas13 systems.Similar to Cas9,Cas12a can target dsDNA and realize cleavage by recognizing PAM sequences(5′-TTTN-3′).It is found that Cas12a not only has thiscis-cleavage activity targeting dsDNA,but also can cut any nearby single-stranded DNA(ssDNA),which is named collateral cleavage activity ortrans-cleavage activity.Besides,the collateral cleavage activity can also be activated by ssDNA.Cas14 has a small size compared with other Cas proteins.It can cleave ssDNA without specific PAM sequence with itscis-cleavage activity and it also hastrans-cleavage activity.Different from the Cas proteins mentioned above,Cas13a can cleave specific RNA with the protospacer flanking site(PFS)sequence and thentrans-cleavage for reporter ssRNA(Table 1).The kinetics of the cleavage varies among Cas types and homologs.For a specific Cas protein,itstrans-cleavage activity is also related to sequences of the target sites and experimental conditions[15].
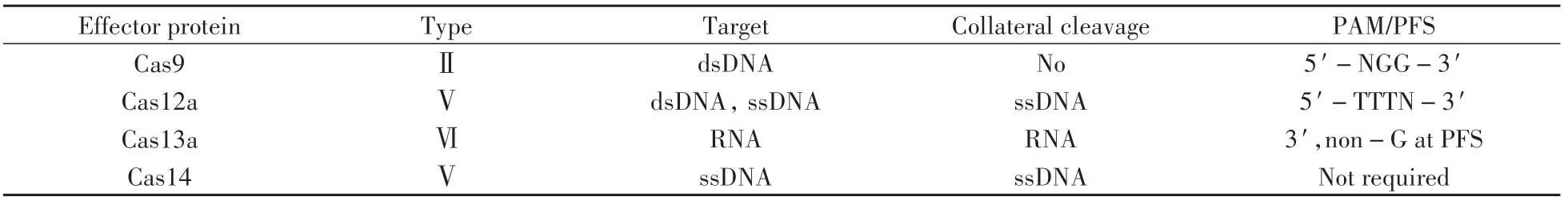
Table 1 Typical CRISPR/Cas systems
2 CRISPR/Cas sensing in molecular diagnosis
Besides the well-recognized gene editing ability,the classⅡCRISPR/Cas system has opened a new way to developing nucleic acid sensing platforms,such as the famous SHERLOCK,DETECTR,and HOLMES[15-17].In such biosensing platform,Cas effector proteins,which possesses unique advantages,such as high detection sensitivity and specificity towards target nucleic acids,can be combined with many different readout methods including,but not limited to fluorescence,colorimetry and electrochemistry,for on-site point-of care testing.For the convenience of the readers,CRISPR/Cas based biosensing platforms and the limits of detection(LOD)are summarized in Table 2.

Table 2 Summary of CRISPR/Cas based biosensing platform for molecular diagnosis
2.1 Fluorescence readout systems
The specific recognition between Cas proteins and the target nucleic acids makes the CRISPR/Cas system,especially CRISPR/Cas12a and CRISPR/Cas13a with thetrans-cleavage activity,as an ideal tool for nucleic acid detection.When the Cas protein bind to the target sequence under the guidance of gRNA,the ternary complex will stimulate its nonspecific cleavage activity and can digest any ssDNA or RNA in the system,which are often labeled with fluorophore and quencher.As a result,the fluorescence resonance energy transfer(FRET)was no longer effective and the substantial fluorescence signal was recovered.Fluorescence readout are most widely used in CRIPSR-powered biosensing system because it can be easily combined with thecis/trans-cleavage activity of CRISPR/Cas systems.
Recently,a CRISPR/Cas-only amplification network(CONAN)for ultrasensitive DNA diagnostics was devised,which took full use of the stringent target recognition,helicase activity,andtrans-cleavage activity of Cas12a(Fig.2)[18].Cas12a and a gRNA for target dsDNA were preassembled(T1),which converted the input of target dsDNA into active Cas12a protein(Cas12a/gRNA-T/DNA complex).A scgRNA,with selfreporting capability,not only output amplified fluorescence signals,but produced multiple active(decaged)gRNA molecules in response to each active Cas12a protein witht rans-cleavage activity.The resulting decaged gRNA,together with the assistant probe,could active the collateral cleavage activity of the other Cas12a protein(T2).In this scenario,each catalytic event of an active Cas12a released a decaged gRNA and sequentially assembled a new active Cas12a protein via T2.As a result,the fluorescence signal was exponentially amplified.This strategy achieves one-step and real-time detection of genomic DNA with attomolar sensitivity and enables the effective detection of hepatitis B virus infection and human bladder cancer-associated single-nucleotide mutation in clinical samples.
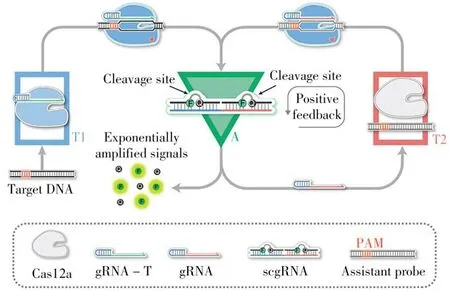
Fig.2 Principle of the CONAN for exponentially amplified detection of DNA using a Cas12a autocatalysis-driven positive feedback circuit[18]
miRNAs with a size of 19-23 nucleotides,play a pivotal role in numerous biological processes by regulating post-transcriptional gene expression,and are becoming biomarkers for many diseases in molecular diagnostics.A CRISPR-CHA method,combining CRISPR/Cas12a with a catalytic hairpin assembly(CHA)circuit,was developed for the sensitive detection of miRNA[19].The CHA circuit was well-designed to convert and amplify each target into multiple programmable DNA duplexes,which served as triggers to initiate thetrans-cleavage activity of CRISPR/Cas12a for further fluorescence signal amplification.Such rational integration resulted in a two-stage amplified detection of mi RNA and was able to decrease the LOD by approximately 6 orders of magnitude compared with the classical CHA assay.By integrating the advantages of CRISPR/Cas12a system and rolling circular amplification(RCA)techniques,Zhang et al.realized highly specific detection method for exosomal miRNAs[20].The dual-specific recognition from miRNA-padlock initiated RCA and CRISPR/Cas12a-triggered specific cleavage not only ensured the high specificity,but also improved the sensitivity.
Due to the PAM sequence not being frequently detected in the genomic sequence,current nucleic acid sensing method may be restrained.To solve this problem,Liu’s group proposed a novel CRISPR-derived microRNA sensing mechanism for miRNA detection[21-22].The target miRNA triggered the nucleic acid amplification process to produce a long single-strand RNA with numerous pre-crRNA repeats,which could be trimmed and recruited by Cas12a actively.As a result,a large number of Cas12a-crRNA complexes were generated and activated by the corresponding dsDNA activators,then the strong fluorescence signal was obtained.This new strategy remarkably suppresses the nonspecific background and relieves the stringent requirement of PAM site in the target sequence.
The outbreak of the novel coronavirus disease(COVID-19)caused by severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)has become a global pandemic.There is an urgent need for a POC,rapid,cost-effective,and selective diagnostic test,that can provide fast and accurate test results[23-24].Pang et al.developed a single-tube assay for SARS-CoV-2 in patient samples,by combining advantages of reverse transcription loop-mediated isothermal amplification(RT-LAMP)with CRISPR/Cas12a[25].The RT-LAMP reagents were added to the sample vial,while CRISPR Cas12a reagents were deposited onto the lid of the vial.Once the amplification finished,the tube was inverted and flicked to mix the detection reagents with the amplicon.Such one-pot,closed-tube reaction is suitable for clinical applications.In a similar way,Fu et al.achieved a rapid and sensitive detection ofSalmonellaand identification of drug-resistantSalmonellaby using the round cap tubes[26].It enabled one-pot assays and reduced aerosol contamination by allowing temporary storage of more Cas12a detection solution than flat cap.
Unlike Cas12a,Cas13a exhibit nonspecific degradation of RNA after specific recognition of target RNA.Based on this principle,Shan et al.achieved to directly detect miRNAs with high specificity and simplicity within 30 min and the LOD as low as 4.5 amol was obtained by this one-step assay[27].Since Cas13a is an RNA-guided RNase and Cas12a is an RNA guided DNase,Tian et al.developed a high-efficient dualgene diagnostic technique based on the orthogonal DNA/RNA collateral cleavage mechanism of Cas12a/Cas13a system[28].Two targets were isothermally amplified by multiplexed reverse transcription-recombinase polymerase amplification(RT-RPA)or RPA.Target 1 DNA amplicons were directly recognized by Cas12acrRNA complexes,while target 2 DNA amplicons were converted into RNA by T7 transcription and recognized by Cas13acrRNA complexes.Orthogonal cleavage of DNA and RNA reporters by target-activated Cas12a/Cas13a induced two colored fluorescence emission,which can be detected using a smartphone.This simple and reliable platform could realize dual-gene detection of SARS-CoV-2 as well as African Swine fever virus(ASFV),and provid an accurate point-of-care screening method for infectious diseases in resourceslimited settings.
The CRISPR/Cas9 system has successfully revolutionized gene editing technology and shows great potential in accurate detection of nucleic acid.Wang et al.developed a Cas9 nickase-based amplification platform(Cas9nAR)to amplify a target fragment from genomic DNA.This strategy exhibited a zeptomolar LOD and a single-base discrimination capability within 60 min at a constant temperature of 37℃,which showed great potential to become a routine assay for the quantitative detection of nucleic acids[29].The ability to unwind dsDNA and generate ssDNA at 37℃is a special feature of Cas9,which facilitates the imaging of target sequence in living cells.By integrating the advantages of powerful CHA amplification and CRISPR/Cas9 system,a novel method was developed for the detection of miRNAs in exosomes with a LOD of 23 fmol/L.More importantly,this method could be applied toin sit uimaging of intracellular miRNAs,that pave the way for applications in bioanalysis and disease diagnostics[30].Extracellular vesicles(EVs)is an endogenous transport system for intercellular transfer of biological cargo,that plays a pivotal role in physiological and pathological processes.Due to the lack of suitable readout systems,the biological effects of EV-mediated RNA transfer are rarely studied.Recently,a highly sensitive CRISPR/Cas9 based reporter system was developed,which made the direct functional study of EV-mediated transfer of small noncoding RNA molecules at single-cell resolution come true[31].Wang et al.constructed a novel RCA-assisted CRISPR/Cas9 cleavage(RACE)method for multiplexed detection of a series of miRNAs in an isothermal manner[32].Extracellular vesicle derived miRNAs are more appealing in disease diagnosis and prognosis evaluation because of the relatively higher abundance and the extraordinary circulating stability.Combining the powerful CRISPR/Cas 9 with RCA,Wang et al.constructed a highly specific detection platform for multiple EV miRNAs detection,which was an attractive tool for multiplexed,specific detection of nucleic acids in point-of-care diagnostics[32].
2.2 Visualized signal readout systems
Visualized biosensing which converts the presence of target molecules into visible color changes has attracted much attention due to its low cost,simplicity,and practicality.Since the signal can be detected by the naked eye,such method does not require expensive or sophisticated instrumentation,which is most suitable for point-of-care diagnosis.
Gold nanoparticles(AuNPs)possess unique optical properties due to the featured localized surface plasmon resonance(LSPR).The binding event between recognition element and the analyte can alter the plasmon resonance absorption of transducer AuNPs,resulting in the color change from red to purple.The color change of AuNPs provides an elegant platform for absorption-based colorimetric detection with AuNPs as signal reporters.Ma et al.developed a smartphone-based visual biosensor for CRISPR/Cas12a powered SARS-CoV-2 diagnostics(Fig.3)[33].Specifically,SARS-CoV-2 N gene were reversely transcribed and amplified using specific primers to obtain dsDNA amplicons.The dsDNA amplicons triggered CRISPR/Cas12a based indiscriminate degradation of a ssDNA that was supposed to link two gold nanoparticles,resulting in the redispersion of gold nanoparticles and thus generating visible color changes.By introducing an extra centrifugation step,this strategy achieved a single-copy resolution for SARS-CoV-2 detection by naked eyes.A semi-quantitative result could also be obtained by using a smartphone App-enabled intube readout[33].A Cas9-based colorimetric method using AuNP as optical probes was reported for DNA monitoring,which showed high specificity in discriminating single-nucleotide mismatch[34].The long ssDNA produced by isothermal amplification could capture the AuNP probes and induce the aggregation of AuNPs,leading to a color change of the mixture from wine red to purple.Recently,a dual signal readout sensing platform for cell-free DNA(cfDNA)was reported based on CRIPSR/Cas12a[35].Specifically,a 20 nm AuNP,which played a crucial role in enhancing the fluorescence signal,was functionalized to an approximately 7 nm long FITC-tagged ssDNA.A 60 nm,complementary ssDNA functionalized could be associated with the 20 nm AuNPs via hybridization and acted as a strong fluorescent quencher of the FITC.Upon activating the CRISPR/Cas12a complex by the target cfDNA,the fluorescent quencher AuNP was separated from the fluorophore,thus metal-enhanced fluorescence was recovered with color changes from purple to red purple.

Fig.3 The trans-cleavage of CRISPR/Cas12a can be utilized to devise fluorescent and colorimetric biosensors for SARS-CoV-2 detection
In addition to AuNPs,some color reactions are also used to construct colorimetric analysis.Gong et al.developed a strand-displacement amplification(SDA)assisted CRISPR/Cas12a method for the colorimetric analysis of Hepatitis B virus(HBV)DNA[36].The SDA was utilized to generate ssDNA that could be partially hybridized with a template DNA to form a trigger DNA.Once the Cas12a was activated,the linker ssDNA was cut with the disassociation of glucose oxidase(GOx)from the surface of magnetic beads(MBs).The released GOx was capable of catalyzing the substrate solution to generate a color change.Similarly,by trimming the G-quadruplex DNAzyme with CRISPR/Cas12a,Chen et al.reported a labelfree colorimetric method for detection ofVi bri o parah aemol yticus,which may cause gastrointestinal disorders in human[37].CRISPR/Cas12a recognized the specific sequence of LAMP products and then destroyed the peroxidase-mimicking activity of the G-quadruplex DNAzyme,which could catalyze the color reaction.
An instrument-free quantification platform was developed on a volumetric bar-chart chip(Fig.4)for visualized detection of single nucleotide polymorphisms(SNPs)[38].Once the specific guide RNA bound to its target DNA and activated the collateral cleavage activity of Cas12a,the platinum nanoparticles(PtNPs)were magnetically separated from the MBs and transferred to catalyze H2O2to generate oxygen.The advancements of the red ink bars resulting from the generated oxygen gas indicated the amounts of target.
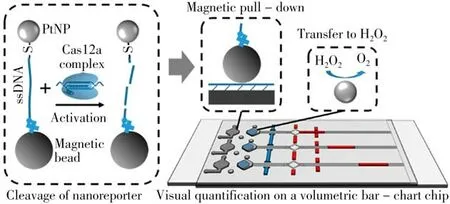
Fig.4 Working principle of the platinum nanoreporterbased CRISPR/Cas12a detection system on the magnetassisted volumetric bar-chart chip[38]
Paper-based lateral-flow assays(LFAs)is one of the most convenient analytical techniques in POC testing with the advantage of being simple,rapid,and cost-effective.Integrating CRISPR/Cas system with LFA improve both the sensitivity and specificity,which makes it more suitable and reliable for clinical usage.By adopting isothermal amplification,CRISPR technology,and new user-friendly readout,several LFAs were developed for the diagnosis of pathogen,such asL i st e r i a monoc yt o gen e s,ASFV and SARS-CoV-2,with a high accuracy when compared to real-time PCR[39-41].Besides the traditional AuNP labels,LFA assay has been improved as a result of the discovery of new labels.Using quantum dots as labels,a simple and low-cost CRISPR/Cas12a-based fluorescence enhanced LFA,combined with recombinase-assisted amplification(RAA),was established to detectS t a p h y l oc oc c us au r eu s[42].Chen et al.designed a dual-mode paper-based strip for the detection of oral squamous cell carcinoma associated miRNA[43].Target miRNA could activate the cascade amplification reaction to generate numerous DNAs,which would further trigger thet r ans-cleavage effect of Cas12a and then generate the naked-eye detectable signal and fluorescent signal.
2.3 Electrochemical signal readout system
Electrochemical signal readout has a unique place in biosensing because of its sensitivity,simplicity and portability. Collaborated with the three-dimensional(3D)DNA walker cascade amplification mechanism,a CRISPR/Cas12a-mediated electrochemiluminescence(ECL)paper-based platform was proposed for the ultrasensitive detection of mi RNA-141[44].As shown in Fig.5,target miRNA-141 could trigger the 3D DNA nanomachine by hybridizing with the protect probe.The released walker probe could combine with the support probe to create restriction recognition sites of Nt.BsmAI nicking endonuclease.The support probe was then specifically sheared,which led to the releasing of the walker probe,thus motivating the 3D DNA walker and releasing a large amount of intermediate DNA.These intermediate DNA could finally act as activator DNA to motivate thetrans-cleavage activity of CRISPR/Cas12a to further achieve efficient annihilation of the ECL signal.Similarly,Zhang et al.proposed the idea of activating CRISPR-Cas12a activity using dsDNA amplified by a 3D DNA walker,and turned it to be a highly sensitive and fast ECL nanosensor system for novel coronavirus target nucleic acids[45].Another ECL biosensor for SARS-CoV-2 based on CRIPSR was constructed rescently[46]. The ECL biosensor employed a DNA tetrahedron structure modified on the electrode surface,and the DNA tetrahedron prongs could form DNA triple-stranded complexes with report DNA.This platform was especially suitable for effective large-scale screening of SARS-CoV-2 in lowresource regions, since the biosensor can be regenerated at pH 10.0.Multiple quantification of biomarkers is attractive in diagnosis,because of its great improvement of accuracy.Bruch et al.developed a CRISPR-powered electrochemical microfluidic biosensor for multiplexed miRNA diagnostics,which can target various RNAs of interest from a single clinical sample using only one effector protein and without changing the sensor or measurement setup[47].
Fig.5 Target conversion principle of 3D DNA walking nanomachine(A).Schematic illustration of the trans-activity/CRISPR/Cas12a-mediated ECL biosensor for the detection of mi RNA-141(B)[44]
3 Conclusions
In the past fifty years,molecular diagnostic technology has made three major transformations:the transformation of report group from radioactive labeling to nonradioactive labeling,the transformation of operation method from manual operation to automatic operation,and the transformation of single target detection to high-throughput detection.CRISPR/Cas system is advanced into a new tool in the field of accurate nucleic acid detection.No matter which signal readout mode is adopted,CRISPR/Cas based biosensing platforms are usually integrated with isothermal nucleic acid amplifications to achieve ultrasensitive detection for target analytes,which can circumvent the thermal cycling process and sophisticated operation of PCR.Most of the sensing strategies have verified their feasibility in complex samples and even clinical samples,holding great promise as a highly efficient tool for molecular diagnosis in POC test.However,there are some shortcomings and more efforts should focus on the following aspects.First,the current proposed biosensor usually contained several steps,including nucleic acids extraction,isothermal amplification and CRISPR/Cas related operations,these multi-step liquid transfer could be further integrated into a one-pot reaction in potential by an automatic process.Second,although the CRISPR/Cas system has been applied for nonnucleic acid targets,not all kinds of biomarkers can be detected because the signal transduction mainly relies on functional nucleic acids such as a specific aptamer.Third,parallel detection of multiple biomarkers and multiple mode signal readout sensing platform should be constructed to improve the accuracy of diagnosis,and eventually transfer such biosensing method from the laboratory benchtop to clinical or practical use.With the further development of molecular diagnosis technology,revolutionary progress in the concept of molecular diagnosis and clinical practical applications is anticipated.
杂志排行
分析测试学报的其它文章
- Research Progress of Hemicyanine Dye for Molecular Imaging
- 碱性磷酸酶的体外检测和体内成像研究进展
- Recent Progress in Nanoscale MOFs for Biological Imaging of Tumors and Tumor Markers
- I-Motif-based Nanosystems for Biomedical Applications:p H Imaging,Drugs Controlled Release and Tumor Theranostics
- A Low-cost,Automated Nucleic Acid Extraction System Converted from the Open-Source Rep Rap 3D Printer
- Research Progress on Analytical Methods for Deciphering Adenosine-to-inosine RNA Editing
