Resistance Evaluation of Indica Rice Varieties with Bacterial Blight Resistance Genes Xa21 and Xa23 under Different Genetic Backgrounds
2022-04-12JianghuiYUJinjiangLILihuaDENGLushuiWENG
Jianghui YU, Jinjiang LI, Lihua DENG, Lushui WENG
Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha 410125, China
Abstract [Objectives] The paper was to evaluate the resistance of indica rice varieties with bacterial blight resistance genes Xa21 and Xa23 under different genetic backgrounds. [Methods] The rice materials with bacterial blight resistance genes Xa21 or Xa23 were used as the donors, and those without the genes were used as the receptors. By back crossing, composite crossing and other methods, combined with molecular marker-assisted selection (MAS), the materials with Xa21 or Xa23 gene, and their sister lines with/without Xa21 or Xa23 gene were inoculated with 7 widely used physiological races of Xanthomonas oryzae, and the resistance of the materials to bacterial blight was analyzed. [Results] The proportions of cumulative resistance (HR, R, MR) of rice materials with resistance gene Xa21 were HNA1-4/PXO86 (84.62%)>YN24 (82.14%)>GD1358 (73.08%)>PXO99 (67.86%)>GDA2 (63.33%) > FuJ (6.67%), indicating that the materials with Xa21 gene had no resistance to bacterial blight induced by strain FuJ, but for other strains, there were still 36.67%-15.38% of the materials susceptible to the disease. The cumulative resistance (HR, R, MR) of the materials with Xa23 gene was over 76%, but there were still 23.02%-15.52% of the materials that had no resistance to the 7 strains. The analysis of variation coefficient of lesion length showed that the resistance of materials with Xa21 or Xa23 genes varied among strains, or the resistance to the same strain varied among materials. Correlation analysis demonstrated that there was significant or extremely significant positive correlation between strain GDA2 or HNA1-4 and other 6 corresponding strains in the materials with Xa21 gene, and there was extremely significant positive correlation between strain YN24, GD1358 or PXO86 and other 6 corresponding strains in the materials with Xa23 gene. Selection of these strains for resistance identification could improve the efficiency of breeding. Analysis of sister lines showed that bacterial blight resistance of Xa21 or Xa23 genes was related to the donor of the materials. Correlation analysis showed that the resistance of the materials with Xa21 gene had extremely significant positive correlation with parents (R=0.572 5**), and the resistance of the materials with Xa23 gene had significant positive correlation with parents (R=0.221 2*). [Conclusions] The study provides the reference for molecular resistance breeding of bacterial blight.
Key words Rice; Bacterial blight; Resistance analysis; Resistance gene
1 Introduction
Bacterial blight is a global bacterial disease, which is known as the "three major diseases" of rice together with rice blast and sheath blight. It is featured by wide range of occurrence, fast prevalence, great harm and high mutability, leading to huge losses in rice production. The yield of infected fields is generally reduced by 10%-20%, or more than 50% in severe cases, or even the whole field fails to harvest[1-4]. Bacterial blight can be effectively controlled by using chemical pesticides or planting resistant varieties. Studies have shown that rice bacterial blight, caused byXanthomonasoryzaepv.oryzae, is a vascular disease, namely, rice vascular infected with the bacteria will produce grayish white lesion, and sugar transporters will transport sucrose to the extracellular space, which results in the proliferation ofX.oryzaein the vascular bundles, thus blocking nutrient transport in the vascular bundles. Meantime, the release of high level of sucrose will interfere with the function of cell membrane, disrupt the signaling pathway of sugar metabolism, and prevent pesticides from directly contacting lesions, resulting in poor effects of pesticides, serious environmental pollution, disruption of ecological balance, and increase of planting cost. Breeding disease-resistant varieties with resistance genes is the most economical and effective way to control bacterial blight and reduce environmental pollution[5-6]. So far, 44 genes (27 dominant genes and 17 recessive genes) against bacterial blight have been reported[7-8], among which 9 genes,Xa1,Xa3/Xa26,xa5,Xa10,xa13,Xa21,Xa23,xa25andXa27, have been cloned, providing excellent genetic resources for breeding disease-resistant varieties[5, 9].Xa4,Xa21andXa23genes are widely used in disease resistance improvement of hybrid rice[10]. It was found thatXa21fromOryzalongistaminataandXa21fromO.rufipogonwere more widely used in practical breeding[11-12]. Bacterial blight resistance genesXa21andXa23are introduced into restorer lines or sterile lines with high yield and good quality but weak disease resistance by conventional breeding and molecular marker-assisted selection (MAS), to develop high quality combination with high resistance and high yield, which has become an important and effective approach for disease resistance breeding in recent years[13-22].
Since the identification of bacterial blight resistance of rice in Hunan Province in 2007, the resistance evaluation of certified rice varieties basically maintained the level of moderate resistance, but the average evaluation grade of bacterial blight resistance of certified rice varieties in 2018 was moderately susceptible (grade 5-6), and the breeding level of bacterial blight resistance decreased[23]. According to the statistics of regional tests or certification of rice in Hunan Province in recent 2 years, among the 120 rice combinations (varieties) that participated in the regional test in 2019, there were only 1 disease-resistant combination (grade 3.0), 34 susceptible combinations (grade 7.0), and 3 highly susceptible combinations to bacterial blight (grade 9.0), with susceptible and highly susceptible combinations accounting for 30.8%, and the rests were moderately susceptible combinations (grade 5.0). Among 136 rice varieties that participated in the regional test in Hunan Province in 2020, there were 75 varieties with bacterial blight resistance above grade 7.0, and the remaining varieties had the resistance of grade 5.0 or above. Among the 103 rice varieties approved by Hunan Province in 2020, only 2 combinations had bacterial blight resistance (grade 3.0), and the remaining varieties had the resistance of grade 5.0 or above. These results suggest the lack of rice varieties against bacterial blight, and it is urgent to breed varieties with resistance to bacterial blight. Meantime, because resistance genes andX.oryzaeare constantly evolving, changes in either could result in the failure of disease-resistant strains. Therefore, it is of great significance to continuously explore or cultivate new disease-resistant materials for bacterial blight resistance breeding, and the breeding of disease-resistant materials for bacterial blight resistance can broaden the resistant spectrum and lengthen the duration of resistance. In this study, taking the materials withXa21andXa23resistance genes against rice bacterial blight as donors, and the materials withoutXa21orXa23genes bred by the research group over the years as receptors, foreground and background breeding was performed through crossbreeding, backcrossing, composite crossing and other methods, combined with molecular marker-assisted selection (MAS), in order to improve the resistance of these materials to bacterial blight or to develop rice lines with wider resistant spectrum and persistent resistance. The resistance of rice materials withXa21orXa23genes against bacterial blight was analyzed by inoculating 7 widely used physiological races ofX.oryzaeinto high-generation stable lines. The resistance of different rice materials to bacterial blight was evaluated by field identification, and the improved resistance lines with excellent comprehensive characters were selected to provide seed resources for bacterial blight resistance breeding and prevention and control of bacterial blight. Meantime, the resistance of the materials withXa21orXa23gene to various physiological races, the correlation between resistance genes and physiological races, and the correlation betweenXa21orXa23gene materials and their sister lines without the genes against bacterial blight were evaluated, in order to provide the reference for molecular resistance breeding of bacterial blight.
2 Materials and methods
2.1 Materials and locationThe experimental materials were 112 rice materials withXa21gene and 117 rice materials withXa23gene, 12 sister lines with/withoutXa21gene, and 26 sister lines with/withoutXa23gene, which were selected and bred by the research group of rice stress tolerance molecular breeding in Institute of Subtropical Agriculture, Chinese Academy of Sciences. These materials had been stabilized beyond the F6generation through hybridization, composite crossing and MAS over the years.
The trial was carried out in 2019 at Changsha Rice Experimental Station, Institute of Subtropical Agriculture, Chinese Academy of Sciences (113°9′ E, 28°9′ N). The area belongs to subtropical monsoon humid climate, with mild climate, abundant rainfall and distinct four seasons. The average annual temperature in the area is 17.2 ℃ and the accumulated temperature is 5 457 ℃.
2.2 MethodsArtificial transplanting cultivation was adopted. The seeds were sown on May 25, and the seedlings were transplanted on June 20, with the row spacing of 16.5 cm×26.4 cm, and the density of 1 seed/hole, 50 clumps each material. Field management was carried out according to conventional cultivation methods. The leaves of individual plants of the test materials were collected 2 weeks after transplanting, and the genomic DNA of rice was extracted by CTAB method[24]. TheXa21andXa23genes of the materials were amplified by 10 μL PCR amplification system consisting of 5 μL of 2×PARMS master mix, 0.15 μL of 10 mmol/L forward primer, 0.15 μL of 10 mmol/L reverse primer, 1.0 μL of template DNA, and 3.7 μL of ddH2O. The selective marker ofXa21gene was the PCR marker pTA248 closely linked[25]; the primer sequence was AgACgCgg AAgggTggTTCCCggA (pTA248-F), CGA TCG CTA TAA CAG CAA AAC (pTA248-R), and the annealing temperature was 60 ℃. The primer sequence of selective marker ofXa23gene was TTGCTCAAGGCTAGGAAAATG (M-Xa23-F), CCCCATCAACGAACTACAGG (M-Xa23-R), and the annealing temperature was 55 ℃. The PCR products were detected by 1.5% agarose gel electrophoresis.
In this study, 7 strains ofX.oryzaewere used for induction, including FuJ and YN24 (dominant pathogenic bacteria in south China rice region), HNA1-4 (resistance identification strain against bacterial blight in the middle and lower reaches of the Yangtze River), GDA2 (representative bacterial blight strain in Guangdong Province), PXO86 and PXO99 (physiological races ofX.oryzaein Philippines), GD1358 (dominant race ofX.oryzaein southern rice region), which were all provided by rice blast identification center of Hunan Plant Protection Institute[26]. The method of leaf cutting inoculation was used for identification. The strains stored at -20 ℃ were activated and revitalized on NA medium, and then cultured at a constant temperature of 28-30 ℃ for 48 h. Finally, the strains were eluted with sterile water to make the bacterial solution uniformly suspended, and the concentration was adjusted to 3×108cfu/mL (OD600=0.5) for inoculation test. During the inoculation, the surgical scissors were sterilized and dipped with well-preparedX.oryzaesuspension. All the materials were inoculated at booting stage. The leaf tips of the test materials were cut off by 2-3 cm, and 5 leaves of each plant were inoculated, 3 plants each material, 3 repeats each treatment. The susceptible rice ‘Jingang 30’ was used as the control and labeled. At about 21 d after inoculation when the state of the test materials tended to be stable, the length of lesions was measured to identify the disease resistance of tested plants.
2.3 Data analysisThe classification standard of bacterial blight was as follows: the average lesion length less than 1 cm, high resistance (HR); 1.1-3.0 cm, resistance (R); 3.1-5.0 cm, moderate resistance (MR); 5.1-12.0 cm, moderate susceptibility (MS); 12.1-20.0 cm, susceptibility (S); greater than 21.0 cm, high susceptibility (HS)[23]. The pathogenicity of rice against 7 strains ofX.oryzaewas analyzed according to the incidence grade of the control material ‘Jingang 30’, and the plant resistance toX.oryzaewas graded according to the standard of resistance grading. Data were statistically calculated by Microsoft Excel 2018, and mean and mean standard deviations were calculated by DPS 15.10 software. The correlation ofXa21andXa23resistance genes among strains, the comparison of sister lines with and without resistance genes, and the correlation of resistance of different materials to bacterial blight strains were analyzed.
3 Results and analysis
3.1 Resistance analysis of the materials withXa21andXa23genes to strains inducing bacterial blight
3.1.1Analysis of resistance frequency. The results (Fig.1) showed that the rice materials with resistance geneXa21had the highest proportion of highly resistant (HR) materials (30.77%) in strain HNA1-4 that induced bacterial blight, while the proportion of highly resistant (HR) materials (3.33%) in strain FuJ was the lowest. The proportion of resistant (R) materials in strain GD1358 was the highest (50%), and the proportion of resistant (R) materials in strain FuJ was the lowest (3.33%). The proportion of moderately resistant (MR) materials in strain PXO99 was the highest (less than 30%), while there was no moderately resistant materials in strain FuJ. The proportions of cumulative resistance (HR, R, MR) among all induced strains were HNA1-4/PXO86 (84.62%) > YN24 (82.14%) > GD1358 (73.08%) > PXO99 (67.86%) > GDA2 (63.33%) > FuJ(6.67%), indicating that the bacterial blight resistance geneXa21had almost no resistance to bacterial blight induced by strain FuJ, but the resistance to bacterial blight induced by strains HNA1-4, PXO86 and YN24 was greater than 80%. However, 36.67%-15.38% of the materials were susceptible to the disease among the 6 strains with high resistance.
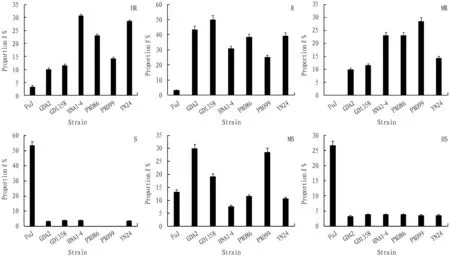
Fig.1 Proportion of Xanthomonas oryzae strains in the materials with Xa21 gene
As shown in Fig.2, among the rice materials with resistance geneXa23, the proportion of HR materials was greater than 50% except HNA2-4; strain YN24 had the highest proportion of 72.18%, followed by strain GD1358, which reached 69.83%. Due to the high proportion of HR materials, the proportion of other resistance materials was relatively small. For R resistant materials, strains HNA1-4 and GDA2 accounted for 27.56% and 26.67%, respectively, and the proportions of other strains were less than 20%. The proportion of MR materials was less than 10% in all strains. However, the cumulative resistance (HR, R, MR) of rice materials among all the induced strains was more than 70%, in which strain GD1358 had the highest resistance and strain FuJ had the lowest resistance. Therefore, 76.98%-84.48% of the materials with bacterial blight resistance geneXa23were resistant to 7 induced strains, indicating thatXa23gene had better resistance to bacterial blight thanXa21gene, with wider antibacterial spectrum, but 23.02%-15.52% of the materials were susceptible to bacterial blight.
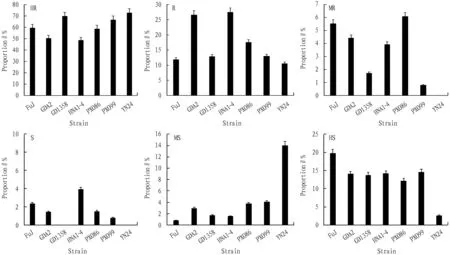
Fig.2 Proportion of Xanthomonas oryzae strains in the materials with Xa23 gene
3.1.2Analysis of lesion length and coefficient of variation. The results (Table 1) showed that the materials with bacterial blight resistance geneXa21had different lesion lengths caused by different inducing strains, and all strains had large coefficients of variation, indicating that the disease resistance varied greatly among materials. Among them, YN24 strain had the largest coefficient of variation, followed by HNA1-4 strain, with the coefficient of variation above 100%, and FuJ strain had the smallest coefficient of variation (41.3%). However, the material withXa23gene had large coefficients of variation among all induced strains, which were all over 140%, and the coefficients of variation of PXO99 and FuJ were over 200%, while the strain PXO86 had the smallest coefficient of variation. In conclusion, the resistance of the same material withXa21orXa23gene to different strains, or the resistance of different materials withXa21orXa23gene to the same strain varied greatly.

Table 1 Lesion length and coefficient of variation of the materials with Xa21 or Xa23 gene induced by 7 strains
3.1.3Correlation analysis of 7 strains inducing bacterial blight of the materials withXa21orXa23gene. Correlation analysis (Table 2) showed that in the materials withXa21gene, there was no significant correlation between strains GD1358 and FuJ, between strains PXO86 and GD1358, between strains FuJ and PXO99, between strains GD1358 and PXO99, and between strains YN24 and FuJ; there were significant (P<0.05) or extremely significant (P<0.01) positive correlations between GDA2 and other 6 strains, and between HNA1-4 and other 6 strains. The results indicated that in the materials withXa21gene, as long as it was resistant to any strain of GDA2 or HNA1-4, it possibly resisted other strains.
Compared with the materials with resistance geneXa21, the materials with resistance geneXa23had higher resistance to bacterial blight. Correlation analysis (Table 3) showed that among the materials withXa23gene, there were extremely significant positive correlations in lesion length among other strains, except that between strains FuJ, GDA2 and HNA1-4, and between strains PXO99 and HNA1-4. Moreover, there were extremely significant positive correlations in lesion length between YN24 and other 6 strains, between GD1358 and other 6 strains, and between PXO86 and other 6 strains, indicating that as long as it was resistant to any strain of YN24, GD1358 or PXO86, it possibly resisted other strains, andXa23gene had broad resistant spectrum to bacterial blight.

Table 2 Correlation analysis of lesion length among 7 strains inducing bacterial blight of the materials with Xa21 gene
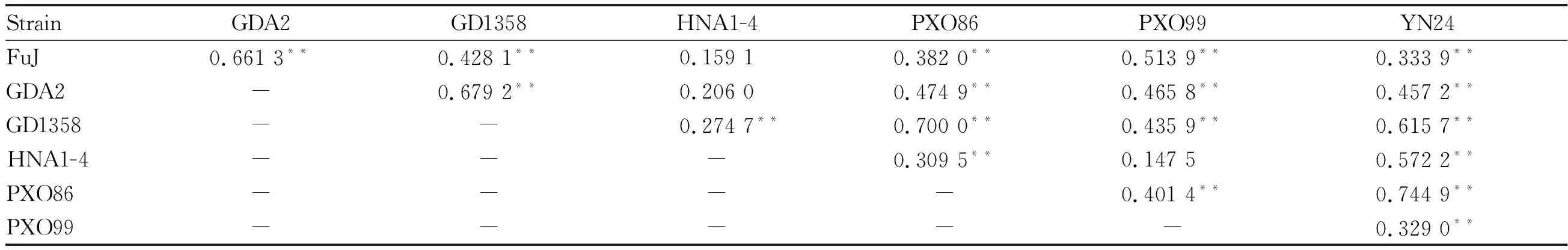
Table 3 Correlation analysis of lesion length among 7 strains inducing bacterial blight of the materials with Xa23 gene
3.2 Resistance analysis of the materials withXa21andXa23gene and their sister lines without the genes
3.2.1Molecular detection. The materials withXa21gene and its corresponding sister lines were detected by M-pTA248marker. The results (Fig.3) showed that lines 01-06 all had the resistance band ofXa21gene, indicating that all the 6 materials contained bacterial blight resistance geneXa21. The sister lines 1-6 all had susceptible bands, suggesting that none of the 6 materials containedXa21gene.

Note: M. DNA marker (DL2000); 01-06, strains with Xa21 gene; 1-6. Corresponding sister lines without Xa21.
The materials withXa23gene and its sister lines were detected by M-Xa23marker. The results (Fig.4) demonstrated that resistance bands (about 200 bp) could be amplified from 07-019 lines, indicating that all 13 lines had bacterial blight resistance geneXa23. The susceptible bands (about 300 bp) could be amplified from 7-19 lines, indicating thatXa23gene was absent in all 13 lines.
3.2.2Comparison ofXa21andXa23genetic materials with gene-free sister lines. After inoculation of bacterial blight strain FuJ, the resistance of the materials withXa21gene and its sister line withoutXa21gene did not change much, and they were still susceptible, proving thatXa21gene had no resistance to bacterial blight strain FuJ (Table 4). Compared with their sister lines withoutXa23gene, 07, 08, 010, 011, 015, 017, and 019 materials withXa23gene had a great change in resistance to bacterial blight, which changed from susceptibility or high susceptibility to resistance or high resistance, while 09, 012, and 013 materials had no change in resistance level, with a resistance frequency of 76.9%. After inoculation of bacterial blight strain GDA2, the resistance of the materials withXa21gene changed significantly compared with their sister lines, and 04 and 05 strains changed from high susceptibility to moderate resistance. The strains 07, 08, 010, 012, 013, 015, 017 and 019 withXa23gene had a higher resistance to bacterial blight, which changed into resistance or high resistance, with the resistance frequency up to 92.3%. After inoculation of bacterial blight strain GD1358, the resistance of 04, 05 and 06 strains withXa21gene changed significantly; the resistance of 08, 012, 015, 017 and 019 strains withXa23gene changed significantly, while 09 and 013 strains were still susceptible, with the resistance frequency up to 84.6%. After inoculation of bacterial blight strain HNA1-4, the resistance of other strains withXa21gene enhanced except for 02 strain, with the resistance frequency up to 83.3%; the resistance of 07, 08, 011, 014, 015 and 019 strains withXa23gene changed greatly, while the resistance of 09, 010, 012 and 013 strains did not improve, with the resistance frequency of 69.2%. After inoculation of bacterial blight strain PXO86, the resistance of other strains withXa21gene enhanced except for 02 strain, with the resistance frequency up to 83.3%; the resistance of other strains withXa23gene enhanced except for 09 strain, with the resistance frequency up to 84.6%. After inoculation of bacterial blight strain PXO99, the resistance levels of other strains withXa21gene increased except for 02 strain, but the resistance frequency was 66.7%; the materials withXa23gene were resistant strains except for 09 and 013 susceptible strains, with the resistance frequency of 84.6%. After inoculation of bacterial blight strain YN24, other materials withXa21gene were resistant or highly resistant except for 02 strain; the materials withXa23gene were resistant strains except for 09 and 013 susceptible strains, with the resistance frequency of 84.6%.

Note: M. DNA marker (DL2000); 07-019, strains with Xa23 gene; 7-19. Corresponding sister lines without Xa23.

Table 4 Pathogenicity of the materials with Xa21 or Xa23 gene and their sister lines without the genes to 7 bacterial blight strains
3.2.3Correlation between resistance ofXa21andXa23gene and materials. Through the correlation analysis of resistance to bacterial blight between the materials withXa21orXa23gene and their sister lines without the genes (Fig.5), it was found that there was extremely significant positive correlation between the resistance of the materials withXa21gene and their sister lines, with the correlation coefficient reaching 0.572 5; there was significant positive correlation between the resistance of the materials withXa23gene and their sister lines, but the correlation coefficient was only 0.221 2.

Fig.5 Correlation between resistance of Xa21 and Xa23 genes and materials
4 Discussion
The indica rice line with bacterial blight resistance geneXa21had the highest resistance to strains HNA1-4 and PXO86 and the lowest resistance to strain FuJ, while the indica rice line with bacterial blight resistance geneXa23had the highest resistance to strain GD1358 and the lowest resistance to strain FuJ, suggesting that strain FuJ had the strongest pathogenicity to the materials withXa21orXa23gene. Wuetal.[27]analyzed the pathogenicity of 12 physiological races ofX.oryzae(including 9 physiological races FuJ, GD414, HEN11, YN1, YN7, YN11, YN18, YN24 and SYb in China and PXO079, PXO145 and PXO099 in the Philippines) in 65 indica rice varieties popularized and applied in different rice regions in China, and found that strain FuJ had the strongest pathogenicity, which could make all tested varieties show susceptible or moderate susceptible response. The results are in agreement with our findings. However, Yangetal.[18]analyzed the resistance of 64 japonica rice varieties to 10 strains ofX.oryzae(YN18, YN1, GD414, HEN11, SYb, YN7, YN11, FuJ, YN24 and Philippine strain PXO99), and found that YN24, PXO99 and YN1 were the most virulent strains, while strain FuJ had weak pathogenicity, which was different from our findings. This was probably because strain FuJ had stronger pathogenicity to indica rice varieties and strain YN24 had stronger pathogenicity to japonica rice varieties. Furthermore, it was found that the rice lines withXa21gene had no resistance to bacterial blight induced by strain FuJ, and 36.67%-15.38% of the materials were susceptible to the other 6 physiological races; 23.02%-15.52% of the materials withXa23gene were susceptible to 7 strains, indicating thatXa23gene had stronger resistance to bacterial blight thanXa21gene. AlthoughXa21gene is highly resistant, it has no resistance to someX.oryzaeraces in Zhejiang and Yunnan in China[13-14], as well as 96% strains in South Korea[15].Xa23gene has attracted the attention of breeders at home and abroad due to its characteristics of broad resistance spectrum, strong resistance, complete dominance and disease resistance during the whole growth period, and has been successively used for the development of molecular markers and breeding research[16-18]. In this study, 7 physiological races ofX.oryzaewere selected, which were widely used in bacterial blight resistance breeding in southern rice region and were highly representative[26]. The correlation analysis of 7 strains inducing bacterial blight of the materials withXa21orXa23gene showed that strains GDA2 and HNA1-4 had significant or extremely significant positive correlation with other 6 corresponding strains in the materials withXa21gene, and materials would possibly resist other bacterial strains as long as they resisted any of these 2 bacterial strains. In the materials withXa23gene, strains YN24, GD1358 and PXO86 were positively correlated with other 6 strains, and materials would possibly resist other bacterial strains as long as they resisted any of these 3 bacterial strains. Because the resistance toX.oryzaestrains is significantly different due to the different resistance genes in the materials, the research results are inconsistent, but such studies have not been reported before.
Zhouetal.[25]transferred bacterial blight resistance geneXa21into an indica rice restorer line ‘9311’ by means of hybridization, backcrossing and self-cross breeding, combined with molecular marker-assisted selection (MAS) breeding, and identified by inoculation with Philippine no. 6 race PXO99; the results showed that the restorer lines of offspring and their hybrid rice combinations had significantly increased resistance to bacterial blight. Studies showed that the restorer lines ‘R8006’ and ‘R1176’ withXa21gene were selected and inoculated by leaf cutting using the main domesticX.oryzaelines III, IV, V and VI, and all showed good resistance[28]. In this study, the resistance analysis of 112 materials withXa21gene and their sister lines with or withoutXa21gene showed that after introduction ofXa21gene, not the resistance to all strains was enhanced, and the materials had no resistance to strain FuJ. Lanetal.[19]obtained ‘YR7009’ and ‘YR7014’ lines withXa21gene through one hybridization, three back-crosses and more than three self-crosses, which had good resistance to 4 bacterial blight strains PXO61, PXO99, ZHE173 and GD1358, but had no resistance to strains FuJ and YN24. Luoetal.[20]introducedXa21into photosensitive genic male sterile line ‘3418S’ and inoculated with 13 representative strains (7 disease prototypes in China and 6 international identification race in Philippines) at tillering stage; the results showed that ‘3418S’ (Xa21) was mostly resistant to the tested strains, but a few showed moderate susceptibility. Therefore, the results of studies on resistance ofXa21gene rice materials to bacterial blight varied among different materials or physiological races. It had a great relation with the background source of breeding materials, while other genes might have antagonistic effects on its expression, and the mechanism should be further analyzed. Our research results suggested that the strain withXa23gene was more affected by genetic background; 7 sister lines had increased resistance, 1 sister line (09 and 9) had no change in resistance, and 5 sister lines had different changes in resistance. Therefore, the introduction ofXa23gene into rice lines in this study could not enhance the resistance of all lines. The single strain inoculation results of previous studies also illustrated that rice materials withXa23gene were not completely resistant to bacterial blight. Yangetal.[29]inoculated 8 sister lines withXa23gene by bacterial blight virulent strain P6, and the results showed that 7 were resistant and 1 was susceptible. Yuetal.[30]carried out field inoculation and identification of bacterial blight on 351 near-isogenic lines withXa23gene through strain P6, and 74 resistant materials were screened, among which 54 were highly resistant materials. Fanetal.[31]obtained 71 and 52Xa23homozygous restorer lines from ‘18113/H705F3’ and ‘18113/H706F3’ strains by hybridization and MAS techniques, respectively, and identified 61 and 44 resistant strains respectively by using Zhe173, a representative strain of IV race ofX.oryzae, for leaf cutting inoculation. Lietal.[32]selected 21 homozygous lines withXa23gene from 160 lines under the same background source and identified the resistance using the identification strain P6, and the results showed that the selected lines were resistant to the disease at seedling stage and booting stage. Although the results vary from study to study, selecting breeding materials by molecular marker-assisted selection closely linked to disease resistance genes can accurately and quickly identify resistance genes or screen resistance sources, and screen materials with good agronomic characters and enhanced resistance from the offspring group, thereby improving the efficiency of breeding. Moreover, multi-generation backcrossing can accelerate the stability of materials and shorten the breeding cycle.
5 Conclusions
By analyzing the resistance of the materials withXa21orXa23resistance genes to 7 strains ofX.oryzae, it was found that the resistance frequency of the materials withXa21gene was more than 60% in 6 strains with high resistance (except FuJ strain), and the resistance frequency of the materials withXa23gene to 7 strains was more than 76%, indicating thatXa23gene had stronger resistance thanXa21gene. Correlation analysis demonstrated that there was significant or extremely significant positive correlation between strain GDA2 or HNA1-4 and other 6 corresponding strains in the materials withXa21gene, and there was extremely significant positive correlation between strain YN24, GD1358 or PXO86 and other 6 corresponding strains in the materials withXa23gene. Selection of these strains for identification could improve the efficiency of breeding. Analysis of sister lines with/without genes showed that the resistance ofXa21gene materials andXa23gene materials had extremely significant or significant positive correlation with parents, respectively, indicating that the resistance of offspring materials was greatly related to the genetic background of parents.
杂志排行
Medicinal Plant的其它文章
- Design and Research of Efficient Adaptive Spray Plant Protection UAV
- Species and Occurrence Regularity of Diseases and Insect Pests Harming Baccaurea ramiflora Lour. in Guangxi
- Effect Analysis of "One Prevention Double Reduction" of Aerial Control in Corn Fields in Daiyue District, Tai’an City
- Development and Application of Green Pest Control Technology in Tea Garden
- AFLP and MSAP Analysis of ‘Lane Late’ Navel Orange and Its Bud Sport Pumpkin-like Navel Orange
- Analysis on the Factors Influencing Pollution-free Vegetable Production: A Case Study of Shenzhen Huantong Farming Products Co. Ltd.
