Unexpected expression of heat-activated transient receptor potential (TRP) channels in winter torpid bats and cold-activated TRP channels in summer active bats
2022-01-27YangYangLiQingYunLvGuanTaoZhengDiLiuJiMaGuiMeiHeLiBiaoZhangShanZhengHaiPengLiYiHsuanPan
Yang-Yang Li, Qing-Yun Lv, Guan-Tao Zheng, Di Liu, Ji Ma, Gui-Mei He, Li-Biao Zhang, Shan Zheng,Hai-Peng Li, Yi-Hsuan Pan,*
1 Key Laboratory of Brain Functional Genomics of Shanghai and Ministry of Education, School of Life Sciences, East China Normal University, Shanghai 200062, China
2 School of Life Science, Department of Biomedical Science, East China Normal University, Shanghai 200062, China
3 Institute of Eco-Chongming, School of Life Sciences, East China Normal University, Shanghai 200062, China
4 Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Sciences, Guangzhou, Guangdong 510260, China
5 CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, Shanghai 200031, China
ABSTRACT The ability to sense temperature changes is crucial for mammalian survival.Mammalian thermal sensing is primarily carried out by thermosensitive transient receptor potential channels (Thermo-TRPs).Some mammals hibernate to survive cold winter conditions,during which time their body temperature fluctuates dramatically.However, the underlying mechanisms by which these mammals regulate thermal responses remain unclear.Using quantitative realtime polymerase chain reaction (qRT-PCR) and the Western blotting, we found that Myotis ricketti bats had high levels of heat-activated TRPs (e.g., TRPV1 and TRPV4) during torpor in winter and coldactivated TRPs (e.g., TRPM8 and TRPC5) during active states in summer.We also found that laboratory mice had high mRNA levels of coldactivated TRPs (e.g., Trpm8 and Trpc5) under relatively hot conditions (i.e., 40 °C).These data suggest that small mammals up-regulate the expression of cold-activated TRPs even under warm or hot conditions.Binding site analysis showed that some homeobox (HOX) transcription factors (TFs)regulate the expression of hot- and cold-activated TRP genes and that some TFs of the Pit-Oct-Unc(POU) family regulate warm-sensitive and coldactivated TRP genes.The dual-luciferase reporter assay results demonstrated that TFs HOXA9,POU3F1, and POU5F1 regulate TRPC5 expression,suggesting that Thermo-TRP genes are regulated bymultiple TFs of the HOX and POU families at different levels.This study provides insights into the adaptive mechanisms underlying thermal sensing used by bats to survive hibernation.
Keywords: Hibernation; Bats; Brain; Thermo-TRPs
INTRODUCTION
Temperature sensing is a complex survival mechanism of living organisms (Castillo et al., 2018; Kashio, 2021).It is accomplished by both the central nervous system and peripheral tissues (Wang & Siemens, 2015).The preopticanterior hypothalamus is the key thermoregulatory region in the vertebrate brain (Cheshire, 2016).It integrates signals from sensory neurons to generate cognitive reflections in response to temperature fluctuations (Tan & McNaughton,2015).Several types of ion channels are involved in the thermosensation of sensory neurons, including thermosensitive leak potassium channels (TREK1, TREK2,and TRAAK), voltage-gated sodium channels (Nav1.7 and Nav1.8), voltage-gated potassium (Kv) channels, and transient receptor potential (TRP) ion channels.Thermosensitive TRP channels (Thermo-TRPs) are non-selective cation channels that allow monovalent and divalent cations (e.g., Ca2+, Mg2+,Na+, and K+) to pass through the cell membrane and sense temperature ranging from noxious cold (<17 °C) to noxious heat (>43 °C) (Castillo et al., 2018; Hoffstaetter et al., 2018;Kashio, 2021).
To date, 28 Thermo-TRPs have been discovered and classified into six subfamilies based on their amino acid sequence homology (Kashio, 2021).Structure similarity in the six transmembrane domains suggests that Thermo-TRPs share common molecular mechanisms of activation(Nadezhdin et al., 2021).Several different stimuli for Thermo-TRP activation have been found, including temperature change, ligand binding, mechanical stimulation, and cytosolic signaling (Hoffstaetter et al., 2018; Kashio, 2021; Wang &Siemens, 2015); however, the mechanisms by which thermo-TRPs respond to these stimuli remain largely unknown(Castillo et al., 2018; Diaz-Franulic et al., 2016; Feng, 2014).Previous studies have revealed the temperature range in which various Thermo-TRPs are activated in mammals(Table 1).For example, TRPV-1, and -2, and -3, which belong to the TRP vanilloid subfamily (TRPV), are activated at temperatures higher than 25 °C (Baez et al., 2014; Palkar et al., 2015).TRPM4 and TRPM5 in the melastatin-like TRP family (TRPM) are activated between 15 °C and 40 °C (Baez et al., 2014; Talavera et al., 2005; Zhong et al., 2016), while TRPM8, TRP-canonical channel 5 (TRPC5), and TRP-ankyrin subtype 1 protein (TRPA1) are activated at temperatures lower than 37 °C (Baez et al., 2014).
As TRPM8, TRPC5, and TRPA1 are activated at low temperatures, they are classified as cold-activated thermal receptors.Others, such as TRPV-1, -2, -3, and -4 and TRPM-2, -4, and -5, are activated at high temperatures and are thus considered as heat-activated sensors.In humans, Thermo-TRPs are classified only as hot-sensitive (e.g., TRPV-1, -2, -3,and -4) and cold-sensitive (e.g., TRPM8, and TRPA1)(Ezquerra-Romano & Ezquerra, 2017) (Table 1).Because TRPV1 and TRPV2 sense noxious heat, they are also considered as hot sensors.TRPV3, TRPV4, and TRPM-2, -4,and -5 have been described as warm sensors (Table 1).The components of Thermo-TRPs are recruited and precisely assembled to sense temperature in a species-, tissue-, and context-specific manner (Wang & Siemens, 2015).The evolution of specialized thermosensors has enabled animals to inhabit various environments with specific temperaturedependent behaviors.
Hibernation is an adaptive strategy used by some animals to survive food shortages and low ambient temperatures.Hibernation involves repeated cycles of torpor and arousal.During torpor, small mammals (e.g., squirrels) can reduce their core body temperature to −2.9 °C (Barnes, 1989),metabolic rate to less than 5% of normal (Carey et al., 2003;Reeder et al., 2012), and heartbeat to approximately 2 beats/min (Dawe & Morrison, 1955).During arousal, they quickly recover their body temperature, metabolic rate, and other physiological conditions to the normal range (Andrews,2019).While mammalian hibernators modify their thermal sensing ability in response to long periods of hibernation(Hoffstaetter et al., 2018; Matos-Cruz et al., 2017), the underlying molecular mechanisms of such adaptation remain largely unknown.
Bats (order Chiroptera) are the only mammals that can fly(Teeling et al., 2005).They are endothermic heterotherms(Lazzeroni et al., 2018) and usually maintain their body temperature around 37 °C.They can enter torpor at ambient temperatures ranging from −10 °C to 21 °C (Webb et al.,2011).Some bats (e.g., Myotis species) can hibernate for more than six months, with body temperatures dropping to 3 °C during torpor and rising rapidly to 35 °C upon arousal(French, 1985; Pan et al., 2013; Stones & Wiebers, 1965).Certain transcription factors (TFs) are implicated in the regulation of thermogenesis and metabolism during hibernation (Han et al., 2015; Morin & Storey, 2009; Nelson et al., 2009; Yin et al., 2016).For instance, peroxisome proliferator-activated receptor α (PPARα) has adaptively evolved to regulate lipid metabolism in response to hibernation(Han et al., 2015).Each TF contains at least one DNA binding domain (DBD) and binds to a specific DNA sequence of its target genes.TFs are grouped into different classes based on their DBDs and typically regulate a group of genes with similar functions (Lambert et al., 2018).
As Thermo-TRPs play pivotal roles in thermal sensing(Baez et al., 2014), we hypothesized that the abundance and expression of these molecules are adjusted and regulated by specific TFs in bats in response to hibernation.To test this, we determined the expression levels of various Thermo-TRPs in Myotis ricketti brains during torpor, 2 h after arousal, 24 h after arousal, and during active states by quantitative real-time polymerase chain reaction (qRT-PCR) and the Western blotting.Bioinformatics analysis was performed to identify TFs that regulate the expression of Thermo-TRPs in hibernators.We also used the dual-luciferase reporter assay to investigate the link between Thermo-TRP genes and their regulatory TFs and temperature-stimulated mice to identify commonstrategies employed by small mammals for thermal sensing.
MATERIALS AND METHODS
Animals
All animal studies were approved by the Animal Ethics Committee of the East China Normal University (Approval No.:AE2012/03001; m20190344).Fifteen adult male M.ricketti bats were captured with hand nets from Fangshan Cave(N39o48‘, E115o42‘) in Beijing, China, in February 2015.Only male bats were used in the study as female bats are often pregnant during winter hibernation.Five torpid bats (n=5) were sacrificed on site while still in torpor.The torpid bats were found hanging upside down at the top of the cave and were motionless.The 10 remaining bats were placed separately into cloth bags and transported to the laboratory within 50 min,where they were maintained at 28 °C.Bats were spontaneously aroused during transportation.Five bats (2 haroused bats, n=5) were sacrificed 2 h after arousal and five bats (24 h-aroused bats, n=5) were sacrificed 24 h after arousal.Active bats (n=5) were captured from the same cave in September 2019.Rectal temperatures were 8–10 °C for torpid, 30–32 °C for 2 h-aroused, and 36–37 °C for 24 haroused and active bats.All bats were sacrificed by cervical dislocation to minimize pain and suffering.
Twenty-four 9-week-old C57BL/6J mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd.(China) and were maintained under a 12 h light-dark cycle at 23 °C.After one week of acclimation, the mice were randomly divided into four groups (n=6 per group) and exposed for 1 h/day to 4 °C,25 °C, 37 °C, and 40 °C, respectively (Huang et al., 2020).These temperatures were chosen as they are in the temperature range whereby mice can adjust their physiological status subsequent to thermosensation by their Thermo-TRPs (Hankenson et al., 2018).The noxious cold temperature (4 °C) is sensed by cold-sensitive TRPM8 and TRPA1; the TRPM4 and TRPM5 sensors are activated at 25 °C; TRPV3 (Liu et al., 2011) and TRPM2 are activated at 37 °C; TRPC5 senses temperatures ranging from 25–37 °C;and the noxious hot temperature (40 °C) is sensed by hotsensitive TRPV1 and TRPV2 (Table 1).No food and water were given during the period of temperature stimulation, and the body weight (Table S2; Supplementary Data 3) of each mouse was recorded daily during the study period.The mice were sacrificed on the sixth day of temperature treatment by cervical dislocation.Their brains were removed, immediately snap frozen in liquid nitrogen, and kept at −80 °C until use.
Binding site analysis of 519 TFs of Thermo-TRP genes
The position frequency matrices (PFMs) of 519 TFs were obtained from the JASPAR database (Fornes et al., 2020) as described previously (Han et al., 2015).The nucleotide sequences 1 kb upstream from the transcription start site,introns, and exons, and 1 kb downstream from the stop codon of each Thermo-TRP gene of 63 mammalian species (15 hibernating and 48 non-hibernating species, Supplementary Table S3) were retrieved from the ENSEMBL (https://www.ensembl.org/index.html) or NCBI databases(https://www.ncbi.nlm.nih.gov/guide/genomes-maps/).These sequences were scanned separately using the matrix-scan tool on the RSAT webserver (http://rsat.sb-roscoff.fr/matrixscan_form.cgi) for TF binding sites.As each TF has its own PFM, the number of TF binding sites in each sequence was calculated at different threshold values (P=10−5to 10−8)(Supplementary Excel File 1) (Liu et al., 2018).The binding sites found in the gene sequences of hibernating, nonhibernating, or both species of mammals were labeled as H,NH, and B, respectively (Supplementary Excel File 2).Labeled TFs are listed in Supplementary Excel File 3, and only TFs with a high threshold value (P≥10−6), labeled with H, were investigated.
The qRT-PCR
Total RNA was extracted from brain tissue using a TransZol Up-Plus RNA Kit (TransGen, Biotech, China) and reverse transcribed to cDNA using a Fastking KT Kit (Tiangen, China).qRT-PCR was carried out using the CFX Connect system(Bio-Rad, USA) and a SYBR Green PCR kit (Qiagen, USA).The primers used are listed in Supplementary Table S1.The relative expression level of each target gene was normalized to that of Gapdh using the comparative 2−ΔΔCTmethod (Livak &Schmittgen, 2001).Three separate reactions and six technical repeats for each gene were performed on samples from 4–5 bats under each state (Supplementary Data 3).Relative expression levels are presented as mean±standard deviation(SD), and statistical significance was determined by one-way analysis of variance (ANOVA) and the post-hoc Holm-Sidak test, with P<0.05 considered significant.

Table 1 List of 10 thermo-TRPs and their temperature thresholds for activation#
Preparation of brain protein for the Western blotting
Protein was extracted from the mashed brain tissue of bats and mice.The tissue (0.1 g) was homogenized in 1 mL of lysis buffer containing 10% glycerol, 2% sodium dodecyl sulfate(SDS), 1.25% β-mercaptoethanol, 3.12 mmol/L ethylenediaminetetraacetic acid (EDTA), 1× protease inhibitor cocktail (Roach), and 25 mmol/L Tris-HCl (pH 6.8) using a Precellys®24 grinder (Bertin Technologies, France).The homogenate was heated at 100 °C for 10 min and clarified by centrifugation at 12 000 g for 15 min at 4 °C.Protein concentration was measured using a Quick Start Bradford protein assay kit (Bio-Rad, USA).Western blotting was carried out as described previously (Han et al., 2015; Huang et al.,2020).Brain protein extracts prepared from mice or rats were used as positive controls, and an equal loading of the sample amount was monitored by Ponceau staining (Supplementary Data 3).Detailed information on protein loading and antibodies used is documented in Supplementary Excel File 4.Antibodies that recognize conserved epitopes of various Thermo-TRPs or TFs across several mammalian species were used.The relative quantity of a protein was calculated by dividing the density value of the protein band on the western blot by the total density value of all protein bands in a lane of a Ponceau-stained blot (Huang et al., 2020; Romero-Calvo et al., 2010; Xue et al., 2018) (Supplementary Data 3).The density value of the protein band background in a Ponceaustained lane was subtracted before being used to determine protein concentration.Three to six independent runs were performed on each protein sample from 3–4 bats under each state.Relative protein abundance is presented as mean±SD.Statistical significance was determined by one-way ANOVA and the post-hoc Holm-Sidak test, with P<0.05 considered significant (Supplementary Data 3).
Dual-luciferase reporter assay
The coding regions of mouse HoxA9, human POU3F1, and human POU5F1, promoter sequence of mouse Trpc5 (2 000 bp before start codon), and 3‘-UTR of human TRPC5 (1 000 bp after stop codon) were chemically synthesized by GenePharma (China).The coding regions of mouse HoxA9,human POU3F1, and human POU5F1 were separately cloned between the HindIII and BamHI sites of the expression vector pcDNA3.1(+)(Invitrogen, USA).The promoter sequence of mouse Trpc5 and 3‘-UTR of human TRPC5 were separately cloned between the XhoI and NotI sites of the reporter vector psi-CHECK-2 (Promega, USA).Plasmids thus constructed were verified by restriction site analysis and sequencing(Supplementary Data 1).Each expression and reporter vector pair was transfected into HEK-293T cells.Luciferase activity of the transfected cells was measured at 24 and 48 h posttransfection using the dual-luciferase reporter assay(Promega, USA).Three replicates were performed on each sample, and the relative luciferase activity was expressed as mean±SD.Statistical significance was determined by one-wayANOVA (with post-hoc Holm-Sidak test), and P<0.05 was considered significant.
Interpretation of statistical results labeled alphabetically
After determining the relative mRNA levels of a specific gene,e.g., TRPV1 (Figure 1A), for each of the four bat states, a bar graph was constructed.In the chart, bars representing the state with the highest gene mRNA abundance were marked"a", and bars representing the states with the second and third highest and lowest abundance were marked "b", "c", and "d",respectively.In addition, significant differences (P<0.05) in values were denoted with different letters (Huang et al., 2020;Liu et al., 2018; Yin et al., 2016).In the small bar chart of TRPV2 (Figure 1A), the bar labeled “bc” indicates that the state had a significantly lower mRNA abundance than the state represented by the bar labeled “a,” but showed no significant difference with the state represented by the bar labeled “b” or “c”.The same principle of interpretation was applied to all bar charts of the Western blotting results.
RESULTS
Expression of Thermo-TRP genes in bats under various states
The mRNA abundance of 10 Thermo-TRPs (heat-activated TRPV-1, -2, -3, and -4 and TRPM-2, -4, and -5; and coldactivated TRPM8, TRPC5, and TRPA1) in the brain of torpid,2 h-aroused, 24 h-aroused, and active M.ricketti bats was determined by qRT-PCR.Results showed that torpid bats had significantly (P≤0.05) higher mRNA abundance of all four heat-activated TRPVs compared to active bats and higher mRNA abundance of heat-activated TRPV-1, -3, and -4 compared to 24 h-aroused bats (P≤0.01) (Figure 1A).The mRNA abundance of warm-sensitive TRPM-2, -4, and -5 and cold-activated TRPM8 and TRPC5 in the bat brain ascendedin the order torpor, 2 h-aroused, 24 h-aroused, and active state (Figure 1B, C).In contrast, the mRNA level of the coldactivated TRPA1 in torpid bats was approximately 1.5-fold (P≤0.01) higher than that in active bats (Figure 1C).These results showed that torpid bats expressed high levels of heatactivated TRPV genes, and 24 h-aroused and active bats expressed high levels of both warm-sensitive TRPM and coldactivated TRP genes (Figure 1D).In addition, torpid bats were found to have a 1.89-fold higher mRNA level of the mitochondrial uncoupling protein 1 (UCP1) (P≤0.01) than active bats (Figure 1E), suggesting that bats use UCP1-dependent thermogenesis during winter hibernation.
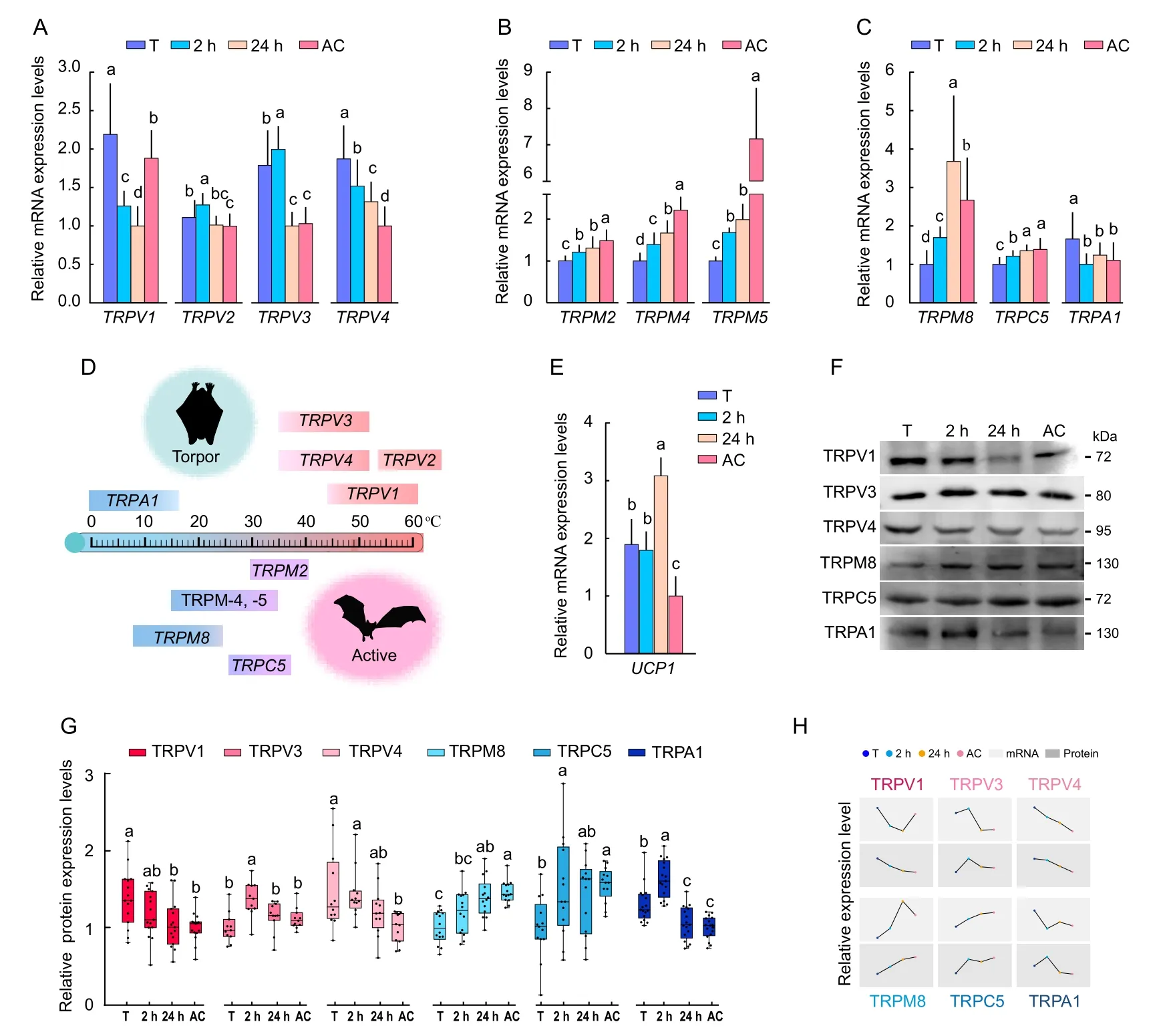
Figure 1 Expression of various Thermo-TRPs in bats under different physiological states
Western blotting was performed to correlate mRNA levels of various Thermo-TRPs with protein expression levels.Results showed that torpid bats had higher protein expression levels of heat-activated TRPV1 (1.5-fold, P≤0.01) and TRPV4 (1.55-fold, P≤0.05) and cold-activated TRPA1 (1.28-fold, P≤0.01)than active bats (Figure 1F, G).The protein levels of heatactivated TRPV3 were similar among bats in all four states but were higher in 2 h-aroused bats than in bats under the other states (Figure 1G).Moreover, active bats showed significantly higher protein expression of cold-activated TRPM8 (1.39-fold,P≤0.05) and TRPC5 (1.41-fold, P≤0.05) than torpid bats(Figure 1F).The Western blotting results generally agreed with the finding (Figure 1D, H) that bats had high mRNAs levels of heat-activated TRPV genes during torpor and of coldactivated TRPM8 and TRPC5 genes under 24 h-aroused and active states.
Identification of possible TFs that regulate Thermo-TRP expression
Binding site analysis was performed to identify TFs that may bind to Thermo-TRP genes (Supplementary Excel Files 3, 5).Results showed that the binding sites of TFs of the FOX family were widely present in different regions of various Thermo-TRP genes (Figure 2A).The binding sites of the HOX TFs were found 1 000 bp upstream of heat-activated TRPV genes and cold-activated TRP genes (Figure 2A), and POU family TFs were found in introns, exons, and 1 000 bp downstream of warm-sensitive TRPM genes and cold-activated TRP genes.Similarly, the binding sites of the TFs of the specificity protein(Sp), transcription factors bind to E2 promotor (E2F), JUND,and erythroblast transformation-specific (ETS) families were found in introns, exons, and 1 000 bp downstream of coldactivated TRP genes (Figure 2A).
To investigate the regulatory roles of TFs on the expression of Thermo-TRP genes, the mRNA expression levels of the following TFs were determined by qRT-PCR: HOXA9,HOXA11, HOXC13, HOXD3, HOXC9, POU1F1, POU2F3,POU3F1, POU3F2, POU4F2, and POU5F1 (Figure 2B, C;Supplementary Excel File 3).Results showed that the mRNA levels of HOXC13, HOXD3, POU2F3, and POU3F1 were 1.46-, 1.42-, 1.17-, and 1.29-fold higher, respectively, in torpid bats than in active bats (Figure 2B, C).In addition, the mRNA levels of HOXA9, POU4F2, and POU5F1 were 1.54-, 1.75-,and 1.63-fold higher, respectively, in active bats than in torpid bats (Figure 2B, C).No significant differences in the mRNA levels of POU1F1 and POU3F2 were found in bats under any state, but the levels of HOXA11 mRNA were higher in 2 haroused bats than in bats under the other states (Figure 2C).
Western blotting showed that the levels of HOXD3,POU3F1, and POU4F2 were 1.44-, 1.61-, and 1.32-fold higher, respectively, in active bats than in torpid bats, while HOXA11, HOXC13, and POU5F1 maintained similar protein expression levels in all bats (Figure 2D, F).The expression patterns of HOCX13, HOXD3, and POU3F1 were similar to those of heat-activated TRPV2, TRPV3, and TRPV4, and that of POU5f1 was similar to its target genes (e.g., TRPM2,TRPM4, TRPM5, TRPM8, and TRPC5) (Figure 2G).
Elevated mRNA levels of Thermo-TRPs and TFs in temperature-stimulated mice
To compare the temperature sensing mechanisms of bats with those of laboratory mice, we determined whether temperature affects the expression of Thermo-TRPs and TFs of the HOX and POU families in the brains of mice by qRT-PCR.Results showed that the mRNA levels of Hoxa9 and Pou3f1 were elevated in 4 °C-treated mice (Figure 3B) and mRNA levels of hot-sensitive Trpv1 were elevated in 25 °C-treated mice(Figure 3A).Significant increases in the mRNA levels of heatactivated Trpv2, Trpv3, Trpv4, Trpm2, Trpm4, and Trpm5(Figure 3A) and in TFs of Hoxc13 and Pou2f3 were observed in 37 °C-treated mice (Figure 3B).In addition, mRNA levels of cold-activated Trpm8 and Trpc5 and Hoxd3 were increased in 40 °C-treated mice (Figure 3A, B).There was a significant drop in body weight (P≤0.05) starting from day 3 in mouse groups under 37 °C and 40 °C treatment compared with mouse groups under 4 °C and 25 °C treatment (Figure 3C),suggesting that the 37 °C and 40 °C treatments adversely affected physiological functions in these mice.
Regulation of TRPC5 expression by HOXA9, POU3F1, and POU5F1
The dual-luciferase reporter assay was performed to determine whether the TFs HOXA9, POU3F1, and POU5F1 regulate the expression of cold-activated TRPC5 (Figure 4A)as it contains many binding sites of these TFs (Supplementary Excel File 6) and exhibited similar increases in mRNA levels in cold and warm surroundings in both bats and mice(Figures 1C, 3A).Results showed that Renilla luciferase activity increased 2.7-fold in cells containing HOXA9(pcDNA3.1-HoxA9-Trpc5 promoter region) (P≤0.05) compared to cells without HOXA9 (pcDNA3.1NC-Trpc5 promoter region)after 24 h (Figure 4B) and luciferase activity was 1.28-fold higher (P≤0.05) in cells containing POU5F1 (pcDNA3.1-POU5F1-TRPC5 3‘-UTR) than in cells without POU5F1(pcDNA3.1-NC-TRPC5 3‘-UTR) at 48 h (Figure 4C).Similarly,luciferase activity was significantly higher in cells with POU3F1 (pcDNA3.1-POU3F1-TRPC5 3‘-UTR) than in cells without POU3F1 (pcDNA3.1-NC-TRPC5 3‘-UTR) at both 24 and 48 h (Figure 4D).
DISCUSSION
We found that torpid and 2 h-aroused bats had lower rectal temperatures than 24 h-aroused and summer active bats.This is consistent with previous findings that the metabolic strategy used by torpid bats is similar to that of 2 h-aroused bats, and that used by 24 h-aroused bats is similar to that of summeractive bats (Huang et al., 2020; Liu et al., 2018).We also found that all bat groups and all mouse groups had a basal expression level of various Thermo-TRPs (Figures 1A, 3A),suggesting that these animals constantly sense and respond to temperature variations.As the amino acid sequence of each bat and mouse Thermo-TRP gene shared approximately 98% similarity with the corresponding Thermo-TRP of other mammals (Table S4; Supplementary Data 2), it is likely that these animals sense temperature changes the same way as other mammals.

Figure 2 Transcription factors (TFs) that regulate Thermo-TRP gene expression
Compared to torpid bats, 24 h-aroused and active bats had a higher abundance of cold-activated TRPs (i.e., TRPM8 and TRPC5) (Figure 1A, H), suggesting that summer active bats have an increased ability to sense cold.In mice, the mRNA expression levels of most heat-activated TRP genes (e.g.,Trpv2, -3, and -4; Trpm-2, -4, and -5) were increased at 37 °C and cold-activated Trpm8, Trpc5, and Trpa1 were elevated at 40 °C (Figure 3A), suggesting common strategies used bymammals in cold sensing under warm or hot conditions.Compared to 24 h-aroused and active bats, torpid bats had a higher abundance of heat-activated TRPs (e.g., TRPV1 and TRPV4) (Figure 1A, H) and cold-activated TRPA1, which senses temperatures <17 °C (Figure 1C) (Karashima et al.,2009), implying that bats have a stronger ability to sense warmth and painful heat (>52 °C) as well as noxious cold(<17 °C) during torpor in winter than during the active state in summer.
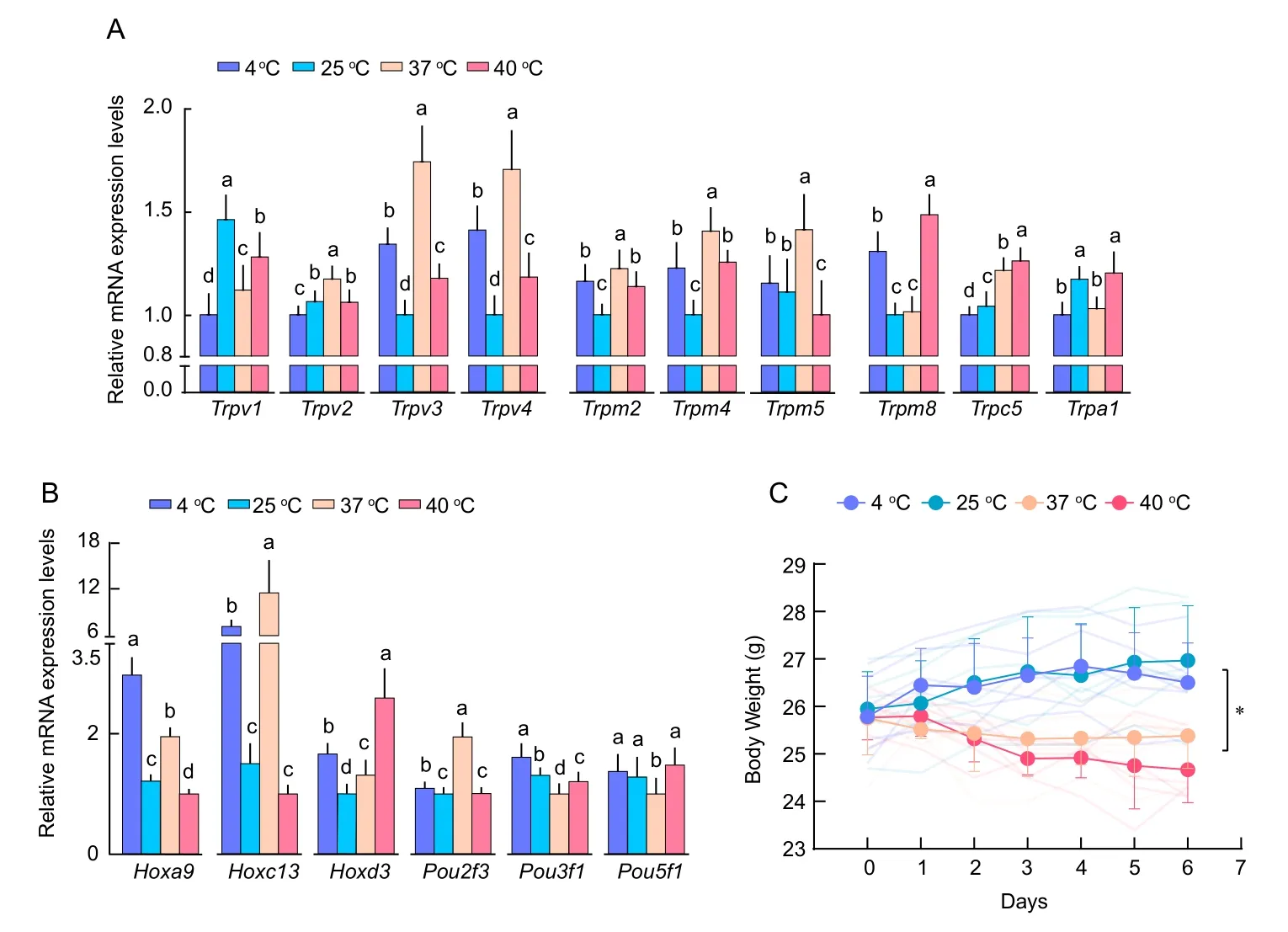
Figure 3 Relative mRNA levels of Thermo-TRPs and TFs in temperature-stimulated mice
Three amino acid residues, i.e., P608, S613, and P623, are known to play pivotal roles in heat activation of human TRPV1(Du et al., 2020).The presence of residues S613 and P623 in the TRPV1 of hibernating bats (i.e., M.brandtii, M.davidii, and M.lucifugus) also suggests a role in heat activation of TRPV1 in these animals.In contrast, two amino acid residues (P613 and S623) in TRPV1 of non-hibernating fruit bats Carollia brevicauda (Du et al., 2020) have a lower threshold temperature for TRPV1 activation (Supplementary Table S5).These findings strongly indicate the role of S613 and P623 in TRPV1 heat sensing in hibernating bats.
The ability of TRPA1 to sense noxious cold has been reported repeatedly, but the classification of TRPA1 as a cold sensor remains controversial (Talavera et al., 2020).However,the presence of amino acid residues S250, M258, and D261 in the ankyrin repeats of TRPA1 in mice and several hibernating bat species (e.g., M.brandtii, M.davidii, and M.lucifugus)suggests that these amino acid residues are important for cold sensing (Supplementary Table S5 and Data 2).This possibility has been confirmed as changing S250, M258, and D261 to N250, L258, and G261 converts TRPA1 from a cold-sensitive to a warm-sensitive sensor (Jabba et al., 2014).
In this study, compared with torpid bats, 24 h-aroused and active bats had higher levels of various warm-sensitive and cold-activated TRPs (Figure 1B, C; Table 1), including TRPM-2, -4, -5, -8, and TRPC5.It is possible that bats have a stronger ability to sense noxious cold (<10 °C) and temperatures ranging from 15 °C to 42 °C (innocuous cold to warm) (Hoffstaetter et al., 2018) in 24 h-aroused and active states than in the torpid state.We found that six amino acid residues, i.e., Y726, S762, S819, S927, H946, and N947, in TRPM8 of hibernating bats are almost identical to those of TRPM8 in humans, mice, and rats, but are very different from those of thirteen-lined ground squirrels (Ictidomys tridecemlineatus) and Syrian hamsters (Mesocricetus auratus)(Supplementary Table S5).These residues have prominent roles in TRPM8 cold sensing (Matos-Cruz et al., 2017) as mutating these residues in squirrels and hamsters to those in humans transforms TRPM8 from a moderately sensitive to a very sensitive cold-sensor (Matos-Cruz et al., 2017).

Figure 4 Regulation of TRPC5 expression by HOXA9, POU3F1, and POU5F1
Our observation of elevated UCP1 mRNA levels in torpid bats agrees well with the finding of elevated UCP1 expression in hibernating thirteen-lined ground squirrels (Laursen et al.,2015) and great roundleaf bats (Hipposideros armiger) (Wang et al., 2014).Previous studies have shown that UCP1 raises body temperature in small mammals by non-shivering thermogenesis (Argyropoulos & Harper, 2002), which is a physiological function of heat production not associated with muscle shivering.Non-shivering thermogenesis occurs mainlyin brown adipose tissue and, to a lesser extent, in skeletal muscle, liver, brain, and white fat (Himms-Hagen, 1984).As only UCP1 can mediate non-shivering thermogenesis in the cold (Golozoubova et al., 2001) and neuronal UCP1 expression indicates local thermogenesis during hibernation(Laursen et al., 2015), up-regulation of UCP1 expression in torpid bats is likely an adaptive strategy used by bats to protect their brain from cold damage during torpor in winter.
TFs of the HOX, POU, and FOX families are involved in body structure formation (Carnesecchi et al., 2018; Reményi et al., 2002), embryonic development (Malik et al., 2018), and antioxidation (Klotz et al., 2015), but their roles in regulating the expression of Thermo-TRPs are not known.Binding site analysis showed that the binding sites of these TFs were widely present in various Thermo-TRP genes (Figure 2A;Supplementary Excel files 1–3, 5, 6), suggesting that TFs of the HOX family regulate the expression of hot- and coldactivated TRP genes and that TFs of the POU family regulate the expression of some heat-activated and cold-activated TRP genes (Figure 2A).The binding sites of the SP and E2F family TFs were found in introns and exons of cold-activated TRP genes and the binding sites of the E2F, JUND, and ETS family TFs were found downstream of these TRP genes.These results suggest that the expression of each Thermo-TRP gene is regulated by multiple TFs at different levels.
A similar expression pattern was seen in HOCX13, HOXD3,POU3F1, and POU5f1 and their target Thermo-TRP genes(Figure 2G), suggesting functional correlation of these TFs and their gene targets (Van Noort et al., 2003).Nonetheless,as TF protein abundance was not consistent with mRNA abundance under all four states in bats (Figure 2F), it is likely that translational control of TFs is independent of their transcription, as previously seen in several hibernating mammals (Storey & Storey, 2010).Although the mechanisms by which bats uncouple transcription and translation to adapt to hibernation remain to be investigated, the significantchange in the expression levels of HOXC13, HOXD3,POU3F1, and POU5F1 in bats of all four states highlights the importance of these TFs in hibernation.
The reporter assay results suggest that HOXA9 enhanced the expression of TRPC5 at the 24 h time point, and both POU3F1 and POU5F1 enhanced the expression of TRPC5 at the 48 h time point.Furthermore, luciferase activity was decreased at 48 h in cells transfected with HOXA9-Trpc5(Figure 4B).This may be due to degradation of HOXA9 as a result of cullin 4A ubiquitination (Zhang et al., 2003).In cells transfected with POU5F1-Trpc5 3‘-UTR, luciferase activity was not detected until 48 h after transfection (Figure 4C), implying that the partners of POU5F1, such as β-catenin and SOX2,also need to be expressed (Li, 2010; Malik et al., 2018).Nevertheless, the reporter assay showed that HOXA9,POU3F1, and POU5F1 were bound to the regulatory regions of the TRPC5 gene and affected its expression.
One limitation of our study is that we examined the expression of Thermo-TRPs in the whole brain instead of the hypothalamus, which is the brain region that regulates thermal responses.This approach was adopted because of our unsuccessful attempts to dissect the hypothalamus in the field due to technical and logistical limitations.Therefore, the expression patterns of bat Thermo-TRPs in the hypothalamus in response to temperature changes remain to be confirmed.Temperature-dependent activity, temperature sensitivity, and activation threshold of bat thermo-TRPs also remain to be characterized in a physiological context.Understanding the molecular basis of these functions will advance our knowledge on the ability of M.ricketti bats to sense warmth and painful heat (>52 °C) and noxious cold (<17 °C) under various physiological conditions.
As Thermo-TRPs have other functions in addition to temperature sensing (Table 1), determining their abundance in the hypothalamus alone may not reveal their physiological roles in other functions.For instance, heat-activated TRPV1 and cold-sensing TRPA1 play significant roles in pain sensation, inflammation, and organ injury (Talavera et al.,2020).In addition, TRPM8 attenuates pain during inflammation (Almeida et al., 2012) and TRPC5 is associated with fear, anxiety, seizures (Zholos, 2014), and cold-induced pain (Bernal et al., 2021).Previous research has shown that TRPV1 in the dorsal root ganglia of vampire bats (Desmodus rotundus) senses heat, but a short isoform of TRPV1 (TRPV1-S) in the trigeminal ganglia of these bats detects infrared radiation (Gracheva et al., 2011).Furthermore, TRPV1 in hibernating thirteen-lined ground squirrel tolerates noxious heat (>40 °C) better than in many other mammalian species(Laursen et al., 2016).Moreover, mice that lose the cold sensor TRPM8 are unable to sense warmth (Paricio-Montesinos et al., 2020).To identify the species-specific functions of various Thermo-TRPs and their collaborations in temperature sensing, comparative studies of TRPVs among bat hibernators and non-hibernators are warranted.
In conclusion, we found that torpid bats up-regulate the expression of heat-activated TRPVs under cold conditions and that active bats up-regulate the expression of both heat- and cold-activated TRPs under warm conditions (Figure 1;Table 1).Laboratory mice also up-regulate the expression of cold-activated TRPs under relatively high temperature, e.g.,40 °C (Figure 3A).Some TFs of the HOX and POU families,such as HOXA9, POU3F1, and POU5F1, regulate the expression of cold-activated TRPC5 (Figure 4).As most cellular machineries work optimally within a narrow temperature, it is conceivable that precise control of temperature sensing is a survival strategy evolved for small hibernating mammals (e.g., bats and squirrels) to overcome repeated torpor-arousal cycles during hibernation.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.Y.L., Q.Y.L., D.L., and J.M.performed the experiments.S.Z.and G.M.H.collected the data.G.T.Z.and H.L.conducted statistical and bioinformatics analyses.L.B.Z.assisted in field work.H.P.L.and Y.H.P.designed the study, provided experimental materials, and wrote the manuscript.All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Dr.Chao-Hung Lee for editing the manuscript and providing valuable advice.
杂志排行
Zoological Research的其它文章
- Zoological Research shines in the East
- Cenozoic Tethyan changes dominated Eurasian animal evolution and diversity patterns
- Genome-wide association study identifies quantitative trait loci affecting cattle temperament
- Opah (Lampris megalopsis) genome sheds light on the evolution of aquatic endothermy
- Comparative mitogenomic analyses unveil conserved and variable mitogenomic features and phylogeny of Chedrinae fish
- A new species of the gudgeon genus Huigobio Fang,1938 (Cypriniformes: Cyprinidae) from the Yangtze River Basin, southern China
