Network pharmacology-based analysis of effective components and mechanism of Rhizoma coptidis in treating diabetes
2021-12-10QianQianZengJiaWeiCaiYueXuLinLiQiuChenRenSongYue
Qian-Qian Zeng, Jia-Wei Cai, Yue Xu, Lin Li, Qiu Chen✉, Ren-Song Yue✉
1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu 610072, P.R. China
2School of Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, 611130, PR China
ABSTRACT
KEYWORDS: Rhizoma coptidis; Diabetes; Network pharmacology;Target; Traditional Chinese medicine
1. Introduction
In the traditional Chinese medicine (TCM) theory, diabetes is classified as "Wasting-thirst", which is a comprehensive syndrome and mainly characterized by excessive drinking, excessive urination,excessive eating, emaciation, fatigue, and sweet urine; while in modern medicine, diabetes is considered as a common metabolic disease with complex pathogenesis, mainly characterized by chronic hyperglycemia. Currently, it is believed that the occurrence of diabetes is related to a variety of factors, such as defective insulin secretion,impaired biological action of insulin, and inflammatory response. The metabolism of diabetic patients could be affected, causing a series of metabolic disorders such as sugar, protein, fat, water, and electrolyte.In addition, long-term hyperglycemia could lead to chronic damage and dysfunction of various tissues, especially eyes, kidney, heart,blood vessels, and nerves, then result in serious complications, such as diabetic nephropathy, diabetic retinopathy, and diabetic peripheral neuropathy and could seriously threaten the life and health of patients[1-3].
Rhizoma coptidis (R. coptidis) is an important traditional Chinese herbal medicine and has been widely used in the treatment of diabetes for thousands of years. It has remarkable curative effects with few side effects and low treatment cost[4,5]. At the same time, modern pharmacological studies have also found that extract of R. coptidis and its active components can inhibit the activity of glucosidase and disglucidases, interfere with the conversion of carbohydrates into monosaccharides and their absorption in the small intestine, and play an anti-diabetic role[6,7]. A study found that berberine, the alkaloid component of R. coptidis, can significantly inhibit the activity of disaccharidases in the protein kinase in a dependent pathway and reduce the mRNA expression of sucrase isomaltase complex in diabetic rats and normal rats[8]. These studies have proved that R. coptidis can directly control the rise of postprandial blood glucose levels by inhibiting carbohydrate digestion. In this study, network pharmacology was used to clarify the pharmacodynamic basis and mechanism of R.coptidis in the treatment of diabetes, and to provide references for the subsequent drug development of R. coptidis.
2. Materials and methods
2.1. Reagent
Phosphate buffer (PBS, 0.1 mol/L, pH 6.8), dimethyl sulfoxide(DMSO), methanol, starch, glucose and sodium carbonate (NaCO)were purchased from Chengdu Klong Chemical Reagent Factory; 3,5 dinitrosalicylic acidchromogen solutions (DNS) was purchased from Chengdu Riefensi Biotechnology Co.; α-glucosidase, α-amylase,bovine serum albumin (BSA), metformin and 4-nitrophenyl-beta-D-glucopyranoside (PNPG) were purchased from Sigma Company.Dulbecco's modified Eagle medium, fetal bovine serum, penicillinstreptomycin, and trypsin-ethylene diamine tetraacetic acid were purchased from Gibco Company. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) detection kit was purchased from Boster Biological Technology Company. 2-Deoxy-2-[(7-nitro-2,1,3 benzoxadiazol-4-yl) amino]-D-glucose (2-NBDG)was provided by Invitrogen Company. Triglyceride (TG) detection kit was purchased from Nanjing Jiancheng Biological Engineering Research Institute. 3T3-L1 pre-adipocytes were acquired from the Beina Biological Company.
2.2. Small molecule database establishment
The chemical components of R. coptidis were collected by searching the database of systematic pharmacology of traditional Chinese medicine, integrated pharmacology-based research platform of TCM V2.0 (www.tcmip.cn/TCMIP/index.), and integrated database of traditional Chinese medicine. Oral bioavailability (OB)and drug-likeness (DL) were used to screen the active compounds.Compounds with sufficient DL≥0.18 and OB≥30% were selected as candidate active ingredients. The 2D or 3D structures of the candidate compounds in PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ChemSpider (http://www.chemspider.com/) were downloaded and saved as a MOL file format.
2.3. Target library establishment
The MOL files of candidate compounds were uploaded to pharmMapper (http://lilab.ecust.edu.cn/pharmmapper/index.php)online database and the potential targets of candidate compounds were identified by high-throughput screening. These target proteins were combined and the repeated collections of small molecule compounds were deleted. The Online Mendelian Inheritance in Man (https://omim.org/), DisGeNET (http:// www.disgenet.org/),TTD (http://bidd.nus.edu.sg/group/cjttd/), and other databases were searched with "diabetes" as the keyword, and the collection of targets of diseases was constructed by eliminating repeated targets. Finally,the small-molecule target set was combined with the disease target set, and the intersection was taken as the target protein library in this study. The standard names of all protein targets were determined by UniProt (http://www.uniprot.org/).
2.4. Bioinformatic annotation
Functional annotation and pathway enrichment analysis of proteins in the target library, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, were carried out by R language software (3.6.3), and bar charts and bubble charts were drawn. Only the top 20 functions or pathways with the most significant enrichment were shown in the pictures. P<0.05 was considered statistically significant. Finally,the potential molecular mechanism of the anti-diabetic effect of R.coptidis was comprehensively analyzed.
2.5. Protein-protein interaction (PPI) network construction
Proteins from the previously obtained target library were uploaded to the string online database to predict PPI and the study species was restricted to human sources. The analysis results were downloaded and saved in TSV format and imported into Cytoscape software(3.6.0) to construct the PPI network of R. coptidis-targets-diabetes.The CytohHubba plug-in and R language (3.6.3) software were used to analyze the hub genes in PPI.
2.6. Network construction
The R language tool was used to construct the compound-target protein data pair and the data pair was imported into Cytoscape software to construct the drug-molecular-target-disease network diagram (DMTD). In this network diagram, the nodes represent drugs, compounds, target proteins, and diseases; while the edges represent the interrelationships between the nodes. Finally, the importance of each node in the network was evaluated by calculating the three topological feature data: degree, average shortest path length, betweenness centrality, and closeness centrality.
2.7. In vitro studies
2.7.1. Preparation of lyophilized powder of R. coptidis extract(RCE)
R. coptidis was purchased from Neautus Chinese Herbal Pieces Ltd.Co., and identified by Prof. Ren-Song Yue of Chengdu University of TCM (HL20191205). A total of 200 g R. coptidis was added to distilled water at a ratio of 1:10, decocted twice for 1 h every time.The liquid was collected and filtered with gauze and the filtrate was centrifuged (12 000 g, 10 min). The supernatant was taken and freeze dried at a low temperature to obtain the lyophilized powder of R.coptidis.
2.7.2. Experiment on inhibition ofα
-glucosidase activityThe inhibitory effect of RCE on the activity of α-glucosidase was determined based on the method of Pierre et al. with some modifications[9]. Briefly, the RCE was first dissolved in PBS containing 10% DMSO and then diluted to different concentrations(0.75-24 mg/mL) for later use. A certain amount of PBS, 20 µL PNPG (0.5 mmol/L), and 20 µL RCE with different concentrations were added to the 96-well plate in turn, and the mixture system was 100 µL. The mixture was oscillated in a water bath at 37 ℃ for 10 min, added with 10 µL of α-glucosidase (0.2 U/mL), mixed well and reacted in a water bath at 37 ℃ for 20 min. Finally, 150 µL NaCOsolution (0.2 mol/L) was added to stop the reaction and the absorbance (OD) of each hole was measured at 405 nm immediately with a microplate. In this study, drugs and α-glucosidase were not added to the blank group, and the RCE was replaced by equal volume PBS as the negative control. Meanwhile, the blank control was set to exclude the influence of drug color on the experimental results (no α-glucosidase solution was added). Each experiment was repeated three times. According to the following formula, the inhibition rate of α-glucosidase activity was calculated.

α
-amylase activityA total of 0.5 mL of α-amylase (0.686 mg/mL) was added into the test tube, then 0.5 mL of RCE with different concentrations was added, mixed evenly, and placed in a water bath at 37 ℃ for 5 min.A total of 0.5 mL 1% soluble starch solution was added, mixed,and reacted for 5 min. Then 0.5 mL DNS was added immediately to color and stop the reaction. The mixture was boiled in boiling water for 5 min, then cooled down to room temperature for 20 min. The volume was set to 5 mL with PBS and the OD of the solution was measured at 540 nm wavelength with a UV-visible spectrophotometer (UV-2550, Shimadzu, Japan). In this study, 0.5 mL PBS was used as the blank control, and the RCE was replaced by equal volume PBS as the negative control. As the experiment of α-glucosidase, the blank control was set to exclude the influence of drug color on the experimental results (no addition of α-amylase).Each experiment was repeated three times. The inhibition rate (%) of the samples with different concentrations to the alpha-amylase was calculated according to the following formula:

2.7.4. Inhibition experiment of advanced glycation end products (AGEs) generation
Fluorescence spectrophotometry (RF-6000, Shimadzu, Japan) was used to detect the inhibitory effect of RCE on AGEs generation.Extract of RCE at different concentrations was added to 1 mL sterile bovine serum albumin (30 mg/mL) and 1 mL glucose solution (0.5 mol/L), mixed evenly, then sealed and incubated for 14 d at 37 ℃in darkness. Fluorescence absorption (FA) of AGEs (emission wavelength 420 nm, excitation wavelength 350 nm) was determined by fluorescence spectrophotometry at room temperature. In this experiment, no drugs or glucose were added to the blank group and the RCE was replaced by PBS of equal volume as the negative control. As mentioned above, the blank control was set to exclude the influence of drug fluorescence absorption on the experimental results (no glucose was added). Each experiment was repeated three times. According to the following formula, the inhibition rate of samples with different concentrations on AGEs generation was calculated.

2.7.5. Cell viability detection
The 3T3-L1 pre-adipocytes were inoculated into 96-well plates (1×10/well) and cultured in an incubator for 24 h. After 24 h, the new medium was replaced. In addition to the normal group, cells in other groups were added with different concentrations of RCE(0-3.2 mg/mL) and incubated for 48 h. After 48 h, the MTT method was used to determine the effect of RCE on the activity of 3T3-L1 pre-adipocytes. In short, 20 µL MTT was added to each well and incubated for another 4 h. The medium was sucked out carefully in the well. A total of 150 µL DMSO solution was added to the well and then the 96-well plate was placed on a low-speed shaking table to oscillate for 10 min until the microcrystals were completely dissolved. The absorbance value of each well was measured at 490 nm using an enzyme marker and the viability of each group of cells was calculated.

2.7.6. Effects of RCE on adipocyte differentiation
The "cocktail method" is often used to cause the 3T3-L1 preadipocytes to differentiate into mature adipocytes, which can well simulate the process of adipocyte differentiation in vitro. In this study, this method was used to induce cell differentiation, and oil red O was used for specific staining of fat. A single-cell suspension was prepared from 3T3-L1 pre-adipocytes in the logarithmic growth stage, seeded in a 96-well culture plate (1×10/well), and cultured for 24 h in a constant temperature incubator at 37 ℃ and 5% CO. When the confluence of cells reached more than 90%, the treatment to promote differentiation was carried out. In addition to the normal group, the cells in the other groups were cultured in the lipid induction medium [complete medium containing 1µmol/L dexamethasone, 0.5 mmol/L 3-Isobutyl-1-methylxanthine(IBMX), 10 µg/mL insulin, and 200 µmol/L indomethacin]for 2 d,followed by complete medium containing 10 µg/mL insulin for 2 d, then by complete medium (without insulin) for 10 d. At the same time of inducing differentiation, cells in the drug group were treated with different concentrations of RCE (0.1, 0.2, 0.4 mg/mL), and cells in the positive group were treated with metformin (100 µmol/L)[10]. After induction, the cells were fixed with 10% formalin for 10 min, treated with 60% isopropanol for 5 min, completely dried at room temperature, and stained with oil red dye for 30 min. Finally,isopropanol was added to the 96-well plate to dissolve the oil red dye. After the oil red in the fat cells was fully dissolved, an enzyme marker was used to detect the OD value of each group at 510 nm.
2.7.7. Glucose uptake assay
With reference to the method described by Alonso-Castro et al.[11],the effect of RCE on glucose uptake by fat cells was evaluated by 2-deoxyglucose uptake assay. The well-differentiated 3T3-L1 preadipocytes were inoculated into 96-well plates (1×10/well) and placed in an incubator for further culture for 24 h. After the cells were completely adherent to the wall, the cells in the other groups were added with different concentrations of RCE (0.1, 0.2, 0.4 mg/mL) or metformin (100 µmol/L) for 48 h. After treatment, 100µmol/L 2-NBDG was added to each well for incubation for 30 min. Subsequently, the absorption of 2-NBDG was determined at 485 nm/535 nm with a fluorescein marker. The experiments were repeated three times, and the data were expressed by the average of the results of the three experiments.
2.7.8. Detection of intracellular TG
Differentiated 3T3-L1 pre-adipocytes were inoculated into a 6-well plate (1×10/well) and cultured in an incubator for 24 h. After the cells were completely adherent to the wall, RCE was treated with different concentrations (0.1, 0.2, 0.4 mg/mL) for 48 h, and cells in the positive group were treated with metformin (100 µmol/L). After the treatment, the cells were collected and broken by ultrasound. The TG content in cells was detected according to the manufacturer's instructions of the TG detection kit. The experiments were repeated three times and the data were expressed by the average of the results of the three experiments.
2.8. Statistical analysis
All the data in this study were analyzed by SPSS 19.0 software.The results were expressed as mean±SD and the differences between groups were analyzed by one-way ANOVA. P<0.05 indicated that the difference was statistically significant.
3. Results
3.1. Active anti-diabetic compound of R. coptidis
According to database search and literature mining, 352 compounds in R. coptidis were collected. Then, screening was conducted according to the screening criteria of DL≥0.18 and OB≥30%, and finally, 14 small molecule compounds with efficacy were obtained.The information of the compounds is shown in Table 1.
3.2. Diabetic targets of active compounds
Through high-throughput screening, 90 targets of R. coptidis were obtained. A total of 10 221 diabetes targets were collected through database search and literature review. By comparing drug targets with disease targets, 84 overlapping targets were obtained.
3.3. GO analysis of target genes
The GO functions of these target genes were enriched and analyzed by R language software, and the analysis results showed that the diabetic protein genes of active components of R. coptidis were mainly enriched in 81 biological processes. It mainly involved hormone receptor activity, G protein-coupled amine receptor activity,ubiquitin-like protein ligase binding, acetylcholine receptor activity,steroid binding, glutathione binding, G protein-coupled serotonin receptor activity, etc. It indicates that R. coptidis can play an antidiabetic role by participating in a variety of biological processes(Figure 1 & Supplementary Table 1).

Table 1. Information on compounds in Rhizoma coptidis.

Figure 1. Gene Ontology (GO) functions enrichment of overlapping target genes (Top 20). A: Bar chart of GO; B: Bubble chart of GO.
3.4. KEGG pathway enrichment analysis

Figure 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis of overlapping target genes (top 20). A: Bar chart of KEGG; B: Bubble chart of KEGG.
The KEGG pathways of these target genes were enriched and analyzed by R language software, and a total of 80 significant pathways were obtained (P<0.05). As shown in Figure 2 and Supplementary Table 2, the main pathways included AGE-RAGE signaling pathway in diabetic complications, proteoglycans in cancer, hypoxia-inducible factor-1 signaling pathway, tumor necrosis factor (TNF) signaling pathway, PI3K-Akt signaling pathway,endocrine resistance, and a variety of cancer-related pathways (such as prostate cancer, bladder cancer, liver cancer, colorectal cancer,non-small cell lung cancer, etc).
3.5. PPI analysis
In PPI, the nodes represent proteins and the connections between nodes represent the interactions between two proteins. The analysis revealed that IL-6, caspase-3, epidermal growth factor receptor,vascular endothelial growth factor A, MYC, estrogen receptor 1, etc.were key targets of R. coptidis for the treatment of diabetes (Figure 3A&B).
3.6. DMTD
After sorting and analyzing the previously retrieved data through R software, a total of 122 pairs of molecular-target data were obtained and imported into Cytoscape software to obtain DMTD. The results are shown in Figure 4. In this network diagram, there were 94 nodes and 122 edges, including 11 compounds (the targets of the three compounds were not included in the overlapping targets) and 84 targets. The circle represents the target, the square represents the component, and the hexagon represents the drug and disease. The network parameter information of all active compounds is shown in Table 2.

Tabe 2. Network parameter information of all active compounds.
3.7. Anti-diabetic effect of R. coptidis in vitro
As shown in Figure 5A, RCE of different concentrations had a significant inhibitory effect on the activity of α-glucosidase, with the inhibitory rate of high concentration of RCE over 80%. Similarly,RCE showed the same significant inhibitory effect on the activity of α-amylase and the inhibitory effect was dose-dependent (Figure 5B).In addition, 0.75 mg/mL-24 mg/mL RCE significantly inhibited the generation of AGEs in vitro (Figure 5C).
3.8. Cell viability assay
As shown in Figure 6A, RCE of lower than 0.8 mg/mL had little effect on the viability of 3T3-L1 pre-adipocytes. However, as the concentration of RCE gradually increased, the influence on the viability of cells increased, indicating that the low concentration of RCE had no toxic effect on cells. Therefore, in the following experiments, we adopted RCE dosing of 0.1 mg/mL, 0.2 mg/mL and 0.4 mg/mL.

Figure 3. Protein-protein interaction network (PPI) of overlapping target genes. A: The key genes of Rhizoma coptidis in PPI (Cytoscape); B: The key genes of Rhizoma coptidis in PPI (R software).
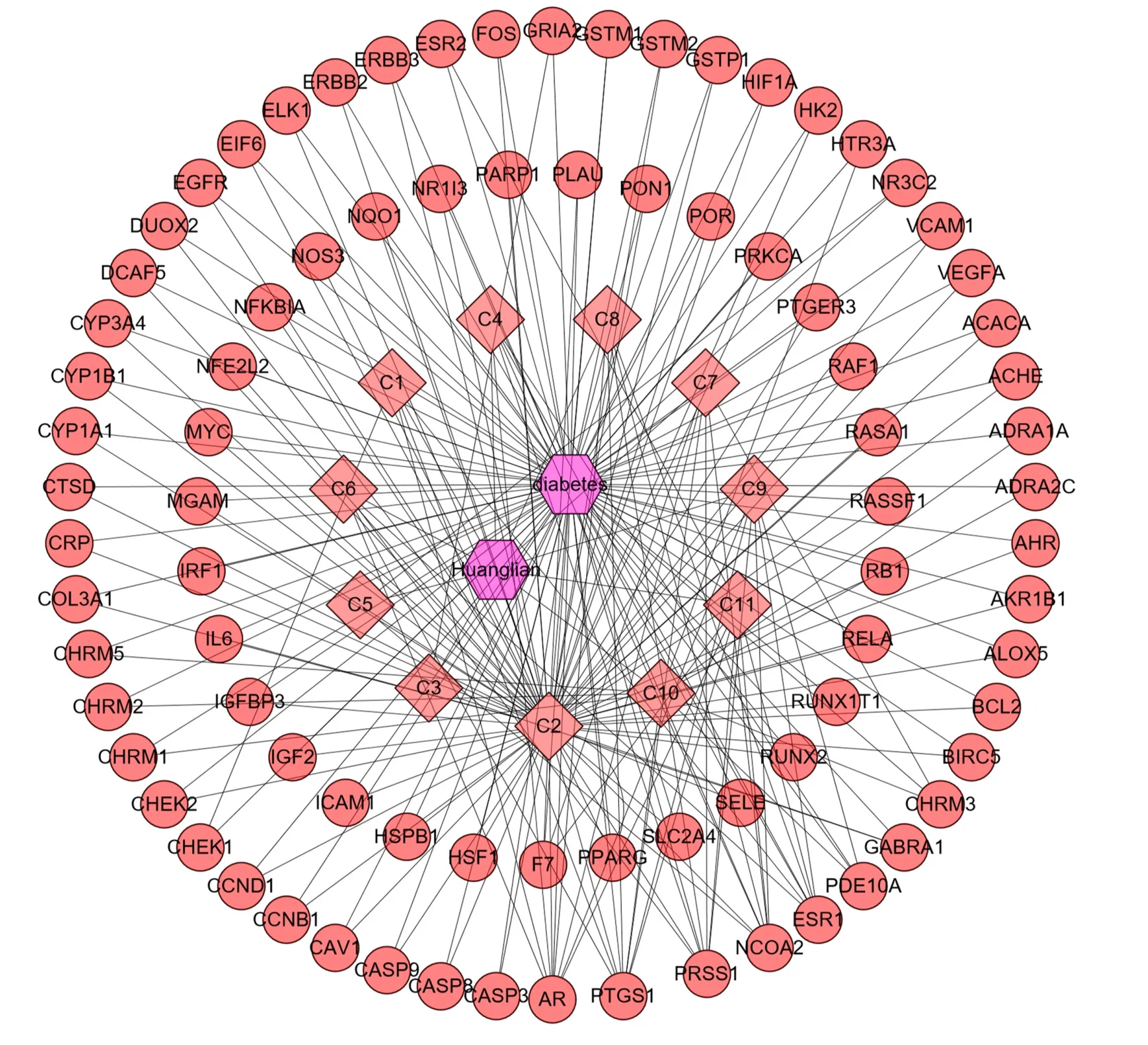
Figure 4. Drug-molecular-target-disease network diagram. The circle represents the target, the square represents the component, and the hexagon represents the drug and disease.
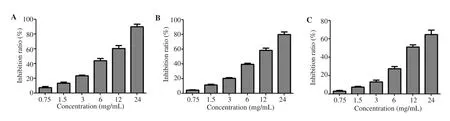
Figure 5. Anti-diabetic effect of Rhizoma coptidis in vitro. A: Experiment on the inhibition of α-glucosidase activity; B: Experiment on the inhibition of α-amylase activity; C: Inhibition experiment of advanced glycation end products generation.
3.9. RCE inhibits adipocyte differentiation
As shown in Figure 6B, compared with the normal group, the OD value of the cells in the control group was significantly increased.It indicated that a large number of lipid droplets were present in the cytoplasm and the pre-adipocytes were successfully induced to differentiate into adipocytes. The results showed that different concentrations of RCE had significant inhibition on adipocyte differentiation, and this inhibition was dose-dependent. Interestingly,the positive drug metformin seemed to have little effect on adipocyte differentiation.

Figure 6. Intervention of extract of Rhizoma coptidis (RCE) on 3T3-L1 pre-adipocytes. A: Cell viability assay; B: effect of RCE on adipocyte differentiation;C: effect of RCE on glucose uptake in cells; D: effect of RCE on lipid metabolism in adipocytes. **P<0.05, ***P<0.01, compared with control.
3.10. RCE promotes glucose uptake by adipocytes
In this study, we investigated the effect of RCE on 2-deoxyglucose uptake by adipocytes. As shown in Figure 6C, the metformin intervention significantly increased glucose uptake in fat cells compared to the control cells. Similarly, RCE also showed a promoting effect in a dose dependent manner. These results suggest that RCE effectively increases glucose uptake in differentiated adipocytes.
3.11. RCE improves lipid metabolism in adipocytes
As shown in Figure 6D, compared with mature fat cells, both metformin and RCE reduced the TG level in cells and the reduction effect of RCE was more significant. It is suggested that RCE can reduce the content of TG in fat cells to improve the disorder of lipid metabolism.
4. Discussion
Diabetes is an incurable metabolic disease characterized by insulin resistance and inadequate insulin secretion[12,13]. At present, the common clinical treatment for diabetes includes insulin injection and oral chemical hypoglycemic agents. However, these treatments have obvious adverse reactions, such as hypoglycemia and gastrointestinal reactions[14]. Herbal medicines have received a lot of attention because of their high efficacy and low side effects in the treatment of diseases[15,16].
TCM believes that the human body is an organic whole, with each tissue and organ communicating with each other structurally,coordinating and serving each other functionally, and also influencing each other in pathology. Therefore, all drugs in TCM play their therapeutic role through multiple channels and targets,which are consistent with the systematic biology thought of network pharmacology. A growing number of studies have found that R.coptidis, an important natural drug, has an obvious anti-diabetes effect[17,18]. In this study, network pharmacology was applied to explore the anti-diabetes effect of R. coptidis and analyze the active ingredients and targets of R. coptidis for the treatment of diabetes. The results showed that 11 chemical components of R.coptidis were closely related to their anti-diabetic effects, including worenine, quercetin, magnograndiolide, epiberberine, corchoroside A, coptisine, berlambine, berberrubine, berberine, (R)-canadine,and palmatine. Through visual analysis, it was found that quercetin,(R)-canadine, and palmatine might be the main active ingredients of R. coptidis for the treatment of diabetes. Currently, studies have found that quercetin has a significant anti-diabetic effect. It can reduce blood glucose by improving the antioxidant status of the pancreas of type 2 diabetic rats and regulating the enzyme activity related to glucose metabolism[19]. In addition, studies have found that quercetin can also improve learning and memory impairment in diabetic rats[20]. Similarly, palmatine therapy has been reported to alleviate neuropathic pain and depressive behavior in diabetes by inhibiting ERK1/2 phosphorylation and TNF-α and IL-1β release in the hippocampus[21]. PPI analysis of the targets of R. coptidis showed that IL-6 was one of the key targets for the action of R. coptidis. In addition, the key targets also include caspase-3, epidermal growth factor receptor, vascular endothelial growth factor A, MYC, estrogen receptor 1, and other targets. These targets have been shown to have an anti-inflammatory effect on diabetes, such as IL-6, which can improve glucose metabolism and play an important role in the treatment of diabetes[22-24]. The consistency of our results with those in previous studies proves the reliability of the results of network pharmacology.
Bioinformatics annotation of overlapping targets showed that these targets were mainly involved in biological processes such as hormone receptor activity, G protein-coupled amine receptor activity, ubiquitin-like protein ligase binding, acetylcholine receptor activity, steroid binding, ubiquitin-protein ligase binding, glutathione binding, G protein-coupled serotonin receptor activity, etc., and involved in pathways such as AGE-RAGE signaling pathway in diabetic complications, proteoglycans in cancer, hypoxia inducible factor-1 signaling pathway, TNF signaling pathway, PI3K-Akt signaling pathway, endocrine resistance, etc. Modern studies have demonstrated that these biological processes and pathways are involved in the development and progression of diabetes in multiple ways[25,26]. Our study indicates that R. coptidis can play an antidiabetic role by participating in multiple biological processes in vivo through multi-component action and multiple targets.
α-Glucosidase plays an important role in the food absorption,food digestion in the intestinal mucosa, and decomposition of starch into glucose[27]. Inhibition of α-glucosidase activity slows the rate at which starch is broken down into glucose and glucose is absorbed in the small intestine. It is worth noting that this process does not generally increase insulin secretion and does not cause hypoglycemia. Therefore, inhibition of α-glucosidase activity is of great significance for the control of glucose homeostasis. α-Amylase can promote the hydrolysis and digestion of carbohydrates in the food. The inhibition of its activity can reduce body sugar absorption,reduce the content of blood glucose and lipid, reduce the synthesis of fat, and induce weight loss[28]. Studies have found that the serum and tissue levels of AGEs in diabetic patients are significantly higher than normal people, and more and more studies have shown that AGEs play an important role in the development of diabetes[29-31]. Natural drugs with the above three activities can be used as a supplement or substitute for diabetes management. The results of this study proved that R. coptidis had significant inhibitory effects on α-glucosidase,α-amylase, and AGEs generation in vitro and preliminarily verified the anti-diabetic effect of R. coptidis. To preliminarily explore the mechanism of action of R. coptidis to treat diabetes, we established the differentiation of the adipose cell model. The results showed that R. coptidis inhibited the differentiation of pre-adipocytes and promoted the glucose uptake of adipocytes. In addition, R. coptidis has a regulatory effect on lipid metabolism disorders in cells by decreasing TG level.
In this study, network pharmacology was used for the first time to explore the scientificity and reliability of R. coptidis in the treatment of diabetes from the perspective of multiple components and multiple targets, and it could provide new ideas for the systematic study of TCM.
Conflict of interest statement
We declare that there is no conflict of interest.
杂志排行
Asian Pacific Journal of Tropical Biomedicine的其它文章
- Identification of climatic and environmental factors associated with incidence of cutaneous leishmaniasis in Central Iran using satellite imagery
- Sang-Yod rice bran hydrolysates alleviate hypertension, endothelial dysfunction,vascular remodeling, and oxidative stress in nitric oxide deficient hypertensive rats
- Immunostimulatory role of rBmHSP60 from filarial parasite Brugia malayi
- Pharmacological aspects of fisetin
