A new genus and species of shrimp (Crustacea:Axiidea:Axiidae) from the Caroline Ridge,Northwest Pacific*
2021-10-12QiKOUGaryPOOREXinzhengLI
Qi KOU ,Gary C.B.POORE ,Xinzheng LI ,
1 Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
2 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
3 Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao),Qingdao 266237, China
4 University of Chinese Academy of Sciences, Beijing 100049, China
5 Museums Victoria, PO Box 666, Melbourne, Vic. 3001, Australia
Abstract A new genus and species of axiid shrimp,Carolinaxius kexuae gen.et sp.nov.is described and illustrated based on a single specimen collected from an unnamed seamount in the Caroline Ridge,Northwest Pacific.Although both chelipeds are missing,the specimen can be distinguished from other axiid genera by a combination of characteristics:narrowly triangular rostrum;median carina and lateral gastric carina each with one prominent tooth;submedian gastric carinae converging posteriorly,with teeth;cornea weakly pigmented,eyestalk with acute distomesial tooth on dorsal surface;male pleopod 1 two-articled;pleopod 2 with appendix interna and appendix masculina;pleopods 3–5 with appendix interna.The molecular phylogeny suggests the new genus is most closely related to another recently described genus living inside hexactinellid sponges on seamounts in the Indian Ocean,Montanaxius Dworschak,2016.However,it differs from Montanaxius in the shape of the rostrum,the arrangement of teeth on the carapace,and the shape of the eyestalk.Besides,the significant molecular differences support the two belonging to different genera.
Keyword:Carolinaxius kexuae gen.et sp.nov.;taxonomy;molecular evidence;deep-sea;seamount
1 INTRODUCTION
In June 2019,an expedition to the Caroline Ridge,Northwest Pacific conducted by Institute of Oceanology,Chinese Academy of Sciences (IOCAS)collected plenty of deep-sea invertebrates by using the remotely operated vehicle (ROV)Faxian.Among the specimens retrieved is an axiid shrimp that proved diffi cult to place in any genus using the keys in the most comprehensive revisions by Sakai and de Saint Laurent (1989),Poore (1994),and Sakai (2011).Nor does it resemble the only more recently described genus from a deep-sea environment (Dworschak,2016) nor any of the deep-sea genera re-diagnosed by Poore (2020).Although the single specimen lacks taxonomically important chelipeds,features of the carapace warrant description of a new genus and species described here.This decision is supported by molecular differences from other axiid genera.
2 MATERIAL AND METHOD
2.1 Collection
This axiid shrimp was found inside the sampling basket installed on ROVFaxianafter the ROV finished a diving on an unnamed seamount in the Caroline Ridge in June 2019.The specimen was immediately fixed and preserved in 75% ethanol after being photographed on board.When the specimens were unloaded and carried to laboratory,fresh 75%ethanol was replaced.The specimens are deposited inthe Marine Biological Museum,Chinese Academy of Sciences (MBMCAS),Qingdao,China.
To resolve the taxonomic status of this novel axiid shrimp,tissue of eight species from six morphologically similar axiid genera and two strahlaxiid species,Neaxiusacanthus(A.Milne-Edwards,1878) andN.trondleiNgoc-Ho,2005,which were used as outgroup,was sampled from collections in Muséum National d’Histoire Naturelle,Paris,France (MNHN),Museums Victoria,Melbourne,Australia (NMV),and University of Florida,Gainesville,USA (UF).The collection locations of all the specimens used in the present study are shown in Table 1.
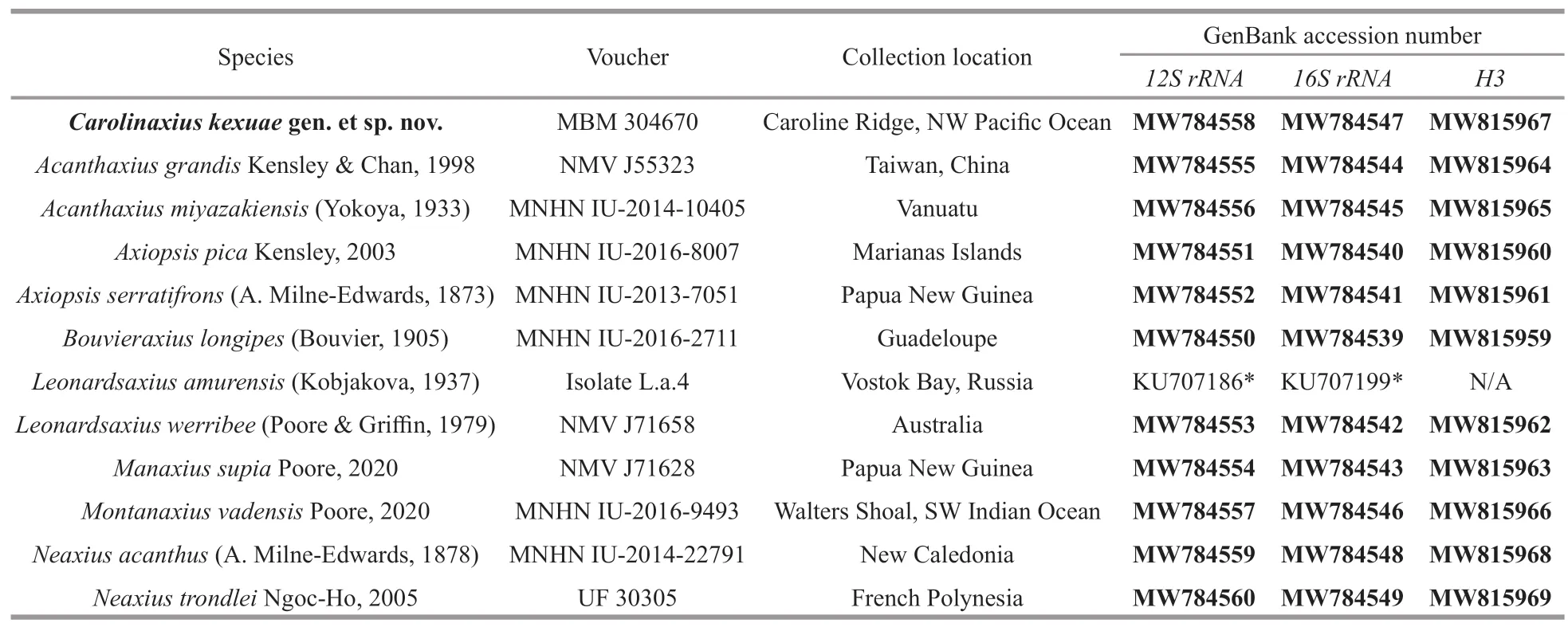
Table 1 Information of specimens used in this study
2.2 Morphology
On board,the axiid shrimp was photographed using a Canon EOS-1D Digital Single Lens Reflex(DSLR) camera.In the laboratory,the specimen was measured and illustrated by using a Zeiss SteREO Discovery V8 stereomicroscope.Carapace length (cl)of the specimen was measured from the tip of the rostrum to the posterior end of the carapace.Total length (tl) of the specimen was measured from the tip of the rostrum to the posterior end of the telson,with body stretched.The generic diagnosis was generated from a DELTA database (Dallwitz,2018) of axiid genera maintained by one of us (Gary C.B.POORE).
2.3 Molecular method
2.3.1 DNA extraction,PCR amplification,and sequencing
Total genomic DNA was extracted from a small piece of muscle tissue of the specimens using a QIAamp DNA Micro Kit (Qiagen,Hilden,Germany)according to the manufacturer’s instructions.Then the DNA was eluted in 50 μL of sterile distilled H2O(RNase free),and stored in refrigerator at -20 °C.Polymerase chain reaction (PCR) amplifications were performed in a reaction mix containing 25 μL of Premix Taq™ (TaKaRa,Japan),5 μL of template DNA,1 μL of each primer (10 mmol/L),and sterile distilled H2O to a total volume of 50 μL.Two mitochondrial ribosomal genes (12Sand16S rRNA)and one nuclear protein coding gene (H3) were amplified using the primers used in Robles et al.(2020).The PCR conditions were as follows:initial denaturation at 94 °C for 5 min,followed by 35−45 cycles of denaturation at 94 °C for 30 s,annealing at 48−53 °C for 40 s,extension at 72 °C for 30 s,and a final extension at 72 °C for 5 min.Before sequencing,the PCR products were purified using the Wizard®SV Gel and PCR Clean-UP System (Promega,Madison,WI,USA).The purified PCR products were sent to Sangon Biotech Co.,Ltd.(Shanghai,China) and bidirectionally sequenced using the same forward and reverse primers for PCR amplification.
2.3.2 Molecular phylogenetic analyses
The forward and reverse sequence fragments were assembled using CONTIG EXPRESS (a component of Vector NTI Suite 6.0,Life Technologies,Carlsbad,CA).The homologous sequences alignment was conducted with MAFFT version 7 webserver (Katoh et al.,2019) using the default parameters,and manually trimmed to the same length.The Kimura’s 2-parameter genetic distances of12Sand16S rRNAgenes were calculated using MEGA 6.06 (Tamura et al.,2013).The highly divergent and poorly aligned regions in the12Sand16S rRNAdatasets were removed using GBlocks v0.91b (Castresana,2000)(GBlocks parameters:minimum length of a block=5;allowed gap positions=with half).Then the trimmed alignments were concatenated into a single dataset consisting of 3 genes (12S/16S/H3) using Sequence Matrix 1.8 (Vaidya et al.,2011).
Phylogenies were inferred based on the concatenated dataset using both maximum likelihood (ML) and Bayesian inference (BI) methods.The concatenated dataset was partitioned by gene and also by codon position for the nuclear protein-coding gene (H3).For ML analysis,the best-fit nucleotide substitution models and optimal partition schemes were selected by ModelFinder (Kalyaanamoorthy et al.,2017)implemented in IQ-TREE 1.6.12 (Nguyen et al.,2015).Then the ML analysis was conducted using IQ-TREE,and the node supports were evaluated by performing ultrafast bootstrap (BP) with 1 000 replicates (Hoang et al.,2018).For BI analysis,the best-fit nucleotide substitution models and optimal partition schemes were determined by PartitionFinder 2.1.1 (Lanfear et al.,2016) with the ‘greedy’ algorithm under the Bayesian information criterion (BIC).The BI tree was reconstructed using MrBayes 3.2.7a (Ronquist et al.,2012).Two independent runs were performed with four Markov Chains for 1 000 000 generations,with sampling every 100 generations.After the first 25%(2 500) trees were discarded as burn-in,the remaining trees were used to construct the 50% majority rule consensus tree and to estimate the posterior probabilities(PP).The effective sample size (ESS) values for all sampled parameters were diagnosed by Tracer 1.7.1(Rambaut et al.,2018) to make sure convergence was reached.The phylogenetic trees were visualized using FigTree 1.4.3 (Rambaut,2016).Finally,all the new sequences were submitted to the GenBank database.
3 RESULT
3.1 Systematics
Order Decapoda Latreille,1802
Infraorder Axiidea de Saint Laurent,1979
Family Axiidae Huxley,1879
Carolinaxius gen.nov.
urn:lsid:zoobank.org:act:399FC78F-4A18-41FEA3D6-78F89D8479B1
Type species:Carolinaxiuskexuaegen.et sp.nov.by monotypy and present designation.
Etymology:From the Caroline Ridge,type locality of the only species,andAxius,type genus of the family.
Diagnosis:Gonochoristic.Carapace smooth.Cervical groove visible laterally over most of distance to anterolateral margin.Rostrum not especially depressed below level of carapace,one-quarter carapace length,narrowly triangular,with few small lateral teeth,continuous with definite lateral carinae.Supraocular spines absent;median carina with 1 tooth;submedian gastric carina converging posteriorly,with 6 or 5 teeth;lateral gastric carina with 1 prominent tooth;postcervical carina absent.Pleonites 2–5 smooth,tergum and pleura continuous;pleura 2–5 rounded.Eyestalk reaching one-third length of rostrum,with acute distomesial tooth on dorsal surface;cornea weakly pigmented.Antenna article 2 distal spine directed anteriorly;scaphocerite simple,straight,almost as long as article 4.Pleurobranchs present above pereopods 2–4;arthrobranchs well developed;epipods on maxillipeds 1–3,pereopods 1–4;podobranchs rudimentary on maxilliped 3–pereopod 3.Maxilliped 3,exopod not clearly bent at base of flagellum.Pereopod 2 dactylus upper margin with few separate setae.Pereopods 3 and 4 dactyli narrow,tapering,with row of spiniform setae near flexor margin.Pereopod 5 dactylus tapering.Male pleopod 1 article 2 triangular,tapering to acute hooked apex bearing subapical seta,mesial margin with blunt appendix interna bearing field of hooks and prominent acute triangular process.Male pleopod 2 endopod with digitiform appendix masculina and appendix interna.Pleopods 3–5 endopod with appendix interna.Uropodal endopod with straight anterior margin obscurely serrate,ending in tooth,distal margin,smooth,continuous with posterior margin;exopod oval,anterior margin barely serrate,with transverse suture,suture ornamented with few simple small spines;article 2 wider than long,without armature.Telson parallel-sided,with semicircular apex,dorsal surface with pairs of teeth,lateral margin serrate,posterior margin convex,with lateral spiniform setae.
Carolinaxiuskexuae gen.et sp.nov.(Figs.1–4)
urn:lsid:zoobank.org:act:1A5076E4-D065-4553-B3C1-2DD298C23D98
Material examined:Male (cl 6.0 mm,tl 19.1 mm),unnamed seamount in the Caroline Ridge,Northwestern Pacific,10°37ʹN,140°05ʹE,796–1 510 m,R/VKexue,collected by crew of R/VKexue,stn FX-Dive 224,12 June 2019,MBM 304670.
Etymology:The specific name of this new species comes from the R/VKexueof Institute of Oceanology,Chinese Academy of Sciences,in recognition of its great contribution to the deep-sea expeditions in the West Pacific Ocean.

Fig.1 Carolinaxius kexuae gen.et sp.nov.Holotype,MBM 304670 (male,cl 6.0 mm,tl 19.1 mm),color in life
Description:Body slender,surface generally glabrous.
Rostrum acutely triangular in dorsal view,0.24 times as long as carapace,2.65 times as long as wide at base,apex acute,reaching to half of antennular peduncle article 3;lateral margins almost straight with 5 (left),4 (right) small,shallow teeth,continuous with lateral carinae;ventral margin unarmed.
Carapace glabrous,with gastric region flat;cervical groove distinct,reaching almost to anteroventral pterygostomial angle;median carina distinct,entire,reaching to cervical groove,armed with 1 strong tooth and 1 shallow tubercle in anterior half;submedian gastric carinae converging posteriorly,armed with longitudinal row of 6 (left),5 (right) prominent independent teeth;lateral carinae short,extending to anterior one-eighth of carapace,widely diverging posteriorly,armed with 1 prominent supraocular tooth(smaller more anterior tooth on left);inferior orbital angle armed with 1 small tooth ;pterygostomial angle strongly produced,unarmed.
Thoracic sternite 6 with median groove separating pair of blunt opposing teeth.Sternite 7 trapezoidal,deeply concave in midline over posterior 1/3,with sharp posterolateral angles.Sternite 8 with subtriangular flap on anterior face at base of pereopod 5.
Pleonites smooth;pleonite 1 0.65 times as long as pleonite 2,pleuron posteroventrally rounded;pleonites 2–5 subequal in dorsal length,pleura posteroventrally rounded,pleonites 3–5 with minute tooth on anteroventral margin;pleonite 6 1.25 times as long as pleonite 5,pleuron ventrally rounded,with a small tooth at midlength,posterolateral projection rounded,posterior margin unarmed.
Eyestalk reaching one-third length of rostrum,trapezoidal in lateral view,with 1 acute distomesial spine on dorsal surface;cornea with numerous pigment granules,division between cornea and eyestalk unclear.
Antennular peduncle reaching to middle of antennal article 5,article 1 with statocyst,with acute stylocerite (on right side);article 2 0.4 times as long as article 1;articles 2,3 subequal in length,1.5 times as long as wide.Antennal peduncle article 1 bearing 1 minute tooth at ventromesial distal angle and 1 tooth on ventrodistal margin;article 2 with blunt distal tooth on ventral surface,distal spine acute,reaching middle of scaphocerite;article 3 with acute ventromesial tooth;article 4 3.5 times as long as wide;article 5 half-length of article 4;scaphocerite slender,acute,reaching distal margin of article 4.
Maxilliped 3 coxa with 1 distomesial spine;basis unarmed;ischium 1.2 times as long as merus,ventral margin with 3 small spines,mesial surface with crista dentata of 19 small teeth;merus armed with 3 spines on ventral margin in distal half,distalmost spine largest,1.3 times as long as carpus;carpus with 1 small subdistal spine on ventral margin,1.3 times as long as propodus;propodus unarmed,1.6 times as long as dactylus;dactylus cylindrical,twice as long as wide;exopod distal two-thirds segmented,reaching middle of second segment of antennal peduncle.
Branchial formula (Table 2).
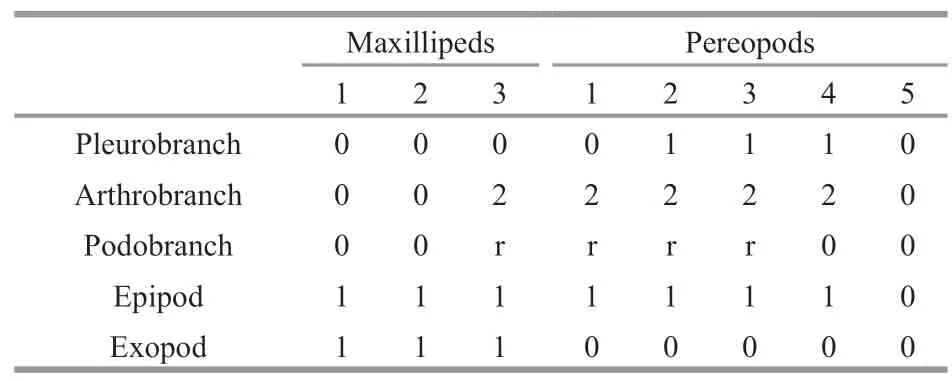
Table 2 Branchial formula of Carolinaxius kexuae gen.et sp.nov.

Table 3 Alignment information and selected DNA substitution model in this study

Table 4 Kimura’s 2-parameter pair-wise genetic distances of 12S rRNA (above diagonal) and 16S rRNA (below diagonal)genes among specimens studied

Fig.2 Carolinaxius kexuae gen.et sp.nov.Holotype,MBM 304670 (male,cl 6.0 mm,tl 19.1 mm)
Pereopods 1 missing.
Pereopod 2 coxa with 2 mesial teeth,1 subdistal tooth on upper margin;basis with 1 distolateral tooth;ischium 2.2 times as long as wide,with 2 prominent teeth on lower margin;merus 3.2 times as long as ischium,upper margin unarmed,lower margin with 2 (left),3 (right) prominent teeth in proximal half,plus 1 subdistal denticle on lateral surface;carpus 0.5 times as long as merus,unarmed;chela setose;palm unarmed,slightly longer than carpus,3.3 times as long as wide;fingers 0.5 times as long as palm,terminating in corneous unguis,cutting edge with row of spinules.
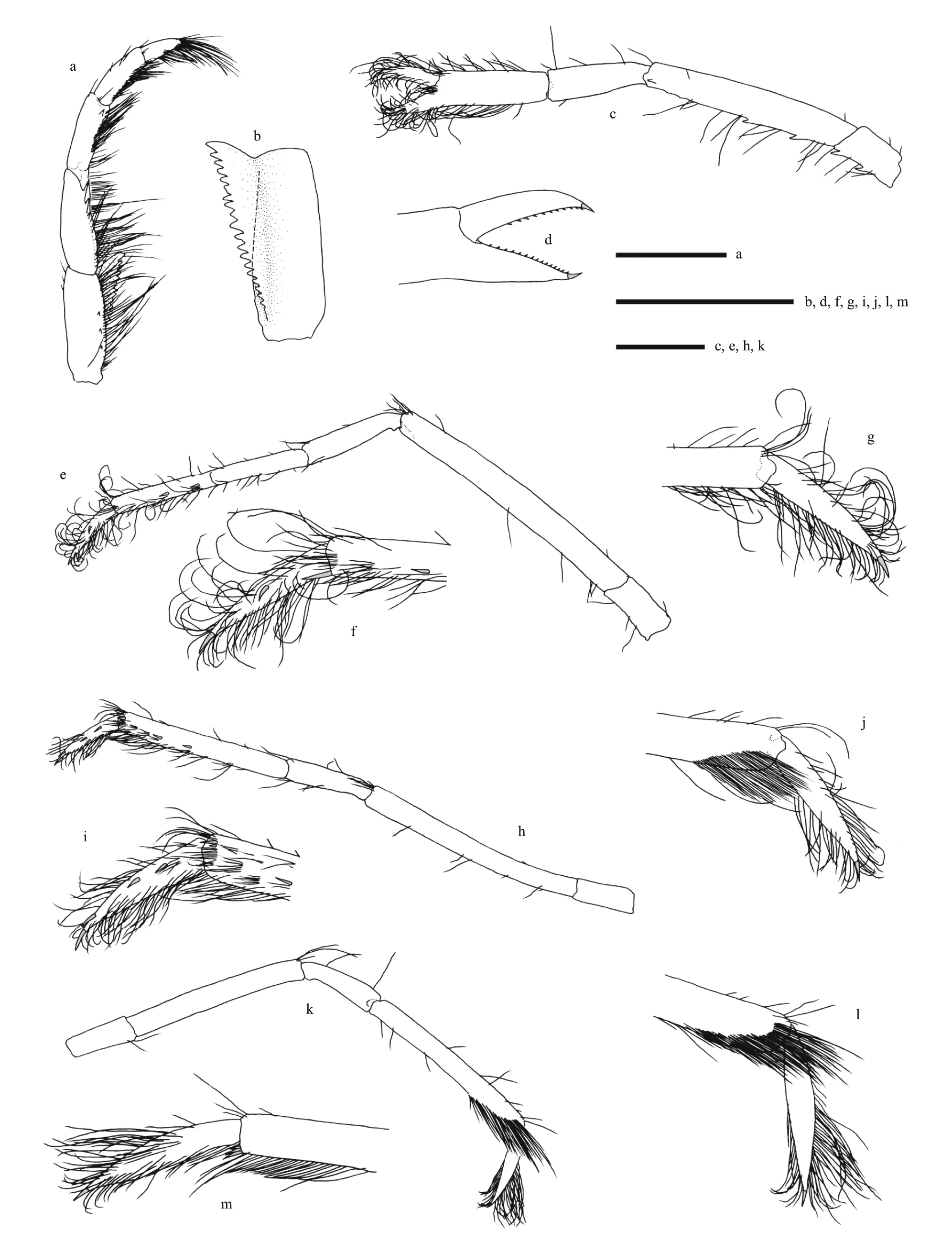
Fig.3 Carolinaxius kexuae gen.et sp.nov.Holotype,MBM 304670 (male,cl 6.0 mm,tl 19.1 mm)
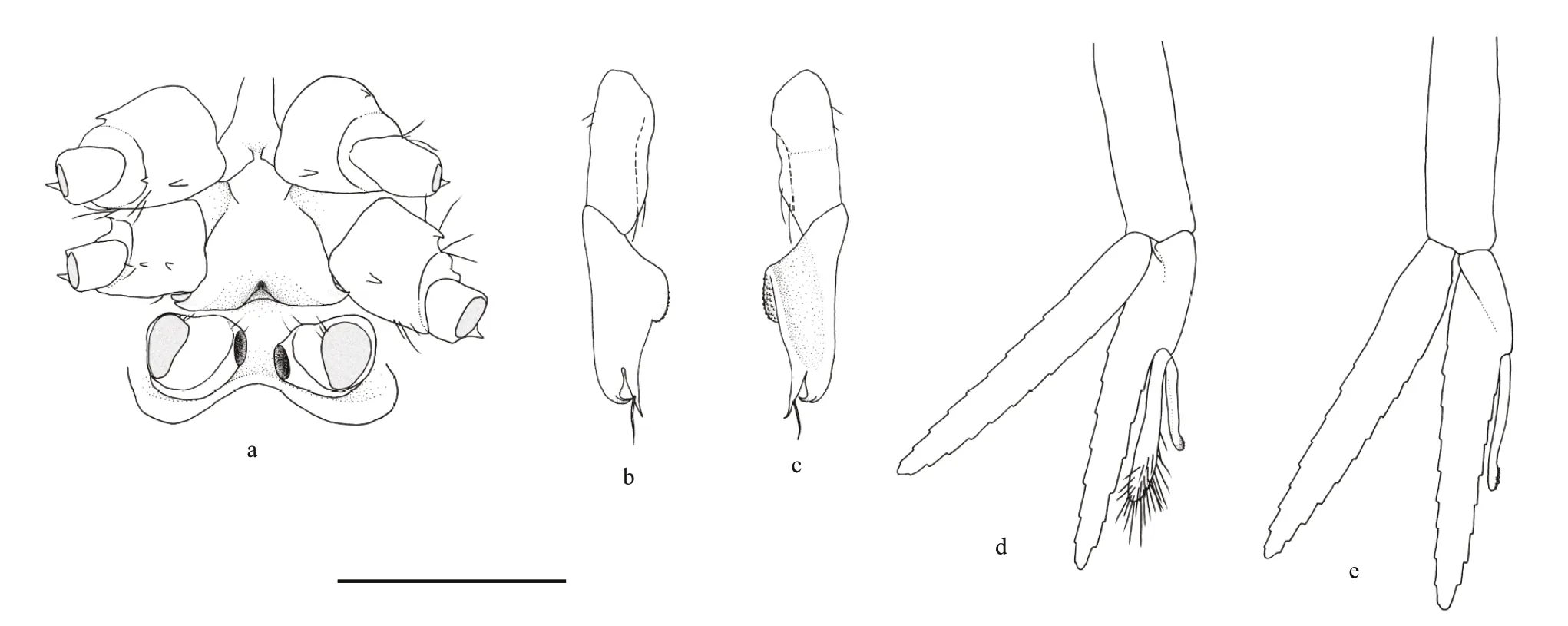
Fig.4 Carolinaxius kexuae gen.et sp.nov.Holotype,MBM 304670 (male,cl 6.0 mm,tl 19.1 mm)
Pereopod 3 coxa with 1 (left),2 (right) mesial teeth,1 subdistal tooth on upper margin;basis with 1 distolateral tooth;ischium twice as long as wide,unarmed;merus 4.5 times as long as ischium,unarmed;carpus 0.4 times as long as merus,unarmed;propodus twice as long as carpus,with sparse setae,lateral surface with 2 (left),3 (right) spinules in distal half,mesial surface unarmed,1 long spinule at lower distal angle;dactylus lanceolate,densely setose,lateral surface with 1 (left),2 (right) spinules,mesial surface unarmed,terminating in corneous unguis.
Pereopod 4 similar to pereopod 3,slightly thinner;propodus lateral surface with 2 spinules in distal half,mesial surface unarmed,distal lower-mesial surface densely setose;dactylus lateral surface with 3 spinules,mesial surface unarmed,terminating in corneous unguis.Pereopod 5 precoxal lobe suboval,coxa with gonopore,coxa to carpus unarmed;propodus distal lower-lateral surface densely setose,with 3 slender spinules at lower distal angle;dactylus lanceolate,densely setose,unarmed,terminating in corneous unguis.
Pleopod 1 uniramous,two-articled;article 2 slightly longer than article 1,tapering to acute hooked apex bearing subapical seta,mesial margin with blunt appendix interna bearing field of hooks and prominent acute triangular process.Pleopod 2 biramous;endopod slightly longer than exopod;appendix masculina 0.4 times as long as endopod,stout,with numerous terminal setae;appendix interna about 0.6 times as long as appendix masculina.Pleopods 3–5 similar,biramous,endopod slightly longer than exopod;with appendix interna,slender,0.4 times aslong as endopod.
Uropod as long as telson;peduncle stout,unarmed;endopod twice as long as wide,anterior margin with 4 (left),3 (right) small teeth over distal two-thirds,distoposterior margin rounded,dorsal surface with longitudinal ridge and row of 6 spines;exopod with almost complete transverse suture,1.7 times as long as wide,slightly shorter than endopod,anterior margin armed with 2 (left),3 (right) small teeth over distal one-third,anterolateral angle with 1 prominent spiniform setae,posterior margin rounded,dorsal surface with longitudinal ridge,unarmed;transverse suture with 3 (left),2 (right) spinules;exopod and endopod both with short simple setae and long plumose setae along posterodistal margins.
Telson subrectangular,1.6 times as long as wide,lateral margin straight,with 4 (left),3 (right) small teeth,posterior margin rounded,with median tooth,posterolateral angle with 3 spiniform setae;dorsal surface armed with 2 pairs of teeth,with scattered long simple setae,posterolateral and posterior margins setose.
Color in life:Body and appendages whitishtranslucent,cornea of eye golden yellow,opaque.
Distribution:Currently only known from the type locality:an unnamed seamount in the Caroline Ridge,Northwestern Pacific,10°37ʹN,140°05ʹE,796–1 510 m.
3.2 Molecular result
A total of 1212S rRNA,1216S rRNA,and 11H3sequences were obtained and included in our analyses.The final concatenated dataset consisted of 1 115 bp(~94.73% of the original 1 177 bp alignment) after the alignments were trimmed with GBlocks (original alignment (12S/16S)=357/476 bp,trimmed alignment(12S/16S)=318/453 bp).The optimum partitioning scheme and the best-fit models for each partitioned subset selected by ModelFinder and PartitionFinder 2 are listed in Table 3.
The Kimura’s 2-parameter pair-wise genetic distances between individuals for the12Sand16S rRNAdatasets are shown in Table 4.For the12S rRNAdataset,the genetic divergences betweenCarolinaxiuskexuaegen.et sp.nov.and other axiid species ranged from 15.5% (Leonardsaxiusamurensis) to 39.7% (Bouvieraxiuslongipes),which were much higher than the intrageneric genetic divergences of three axiid genera (5.4%–11.3%).For the16S rRNAdataset,the genetic divergences betweenCarolinaxiuskexuaegen.et sp.nov.and other axiid species ranged from 20.8% (Montanaxiusvadensis) to 29.7% (Acanthaxiusgrandis),which were also significantly higher than the intrageneric genetic divergences of three axiid genera (5.8%–9.5%).
After 1 million iterations,the average standard deviation of split frequencies in BI analysis reached below 0.004.The phylogenetic trees inferred from both ML and BI analyses were very congruent and generally well supported (Fig.5).In our tree,the ingroup (Axiidae) was recognized as a monophyletic group with high support values (BP=100%,PP=1.00).Amongst the ingroup taxa,Acanthaxius,Axiopsis,andLeonardsaxiuswere recovered as monophyletic with high support (BP ≥98%,PP=1.00).Carolinaxiuskexuaegen.et sp.nov.clustered withMontanaxiusvadensisbut with low support (BP=67%,PP=0.67).However,Carolinaxiusgen.nov.,Montanaxius,andLeonardsaxiusformed a well-supported clade in both analyses (BP=97%,PP=1.00).
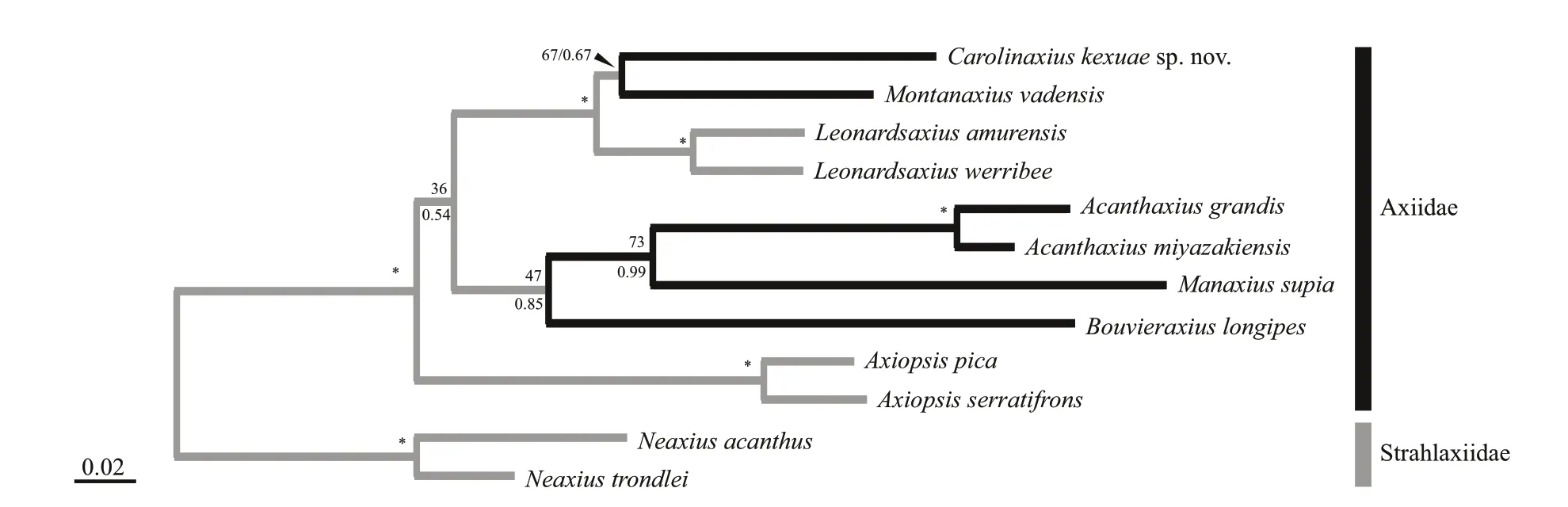
Fig.5 Maximum likelihood tree inferred from the concatenated dataset of 12S rRNA,16S rRNA,and H3 gene sequences
4 DISCUSSION
4.1 Morphological comparison
Some of the genera of the family Axiidae are poorly diagnosed and are probably synonyms (work in progress).Nevertheless,the new species is not a member of the ‘calocaridid’ group of genera because it is gonochoristic and lacks the characteristic pleopod 2 diagnosing this group (Kensley,1989).Most axiid genera have fully pigmented cornea on welldeveloped mobile eyestalks but loss of pigmentation occurs in apparently unrelated genera.Some species of two large deep-water genera associated with sponges,EiconaxiusBate,1888 andSpongiaxiusSakai &de Saint Laurent,1989,have poorly pigmented eyes.Both these genera have a short rostrum and often lack a suture on the uropodal exopod (Poore,2020).Four other monotypic genera have unpigmented cornea.AnophthalmaxiuseccoptodactylusDe Man,1905,has a short simple rostrum,lacks teeth on the median and lateral gastric carinae,and lacks submedian gastric carinae (Sakai,2011).LevantocarishornungaeGalil &Clark,1993,has unpigmented cornea on a cylindrical eyestalk whereas it is flattened in the new species.The two species have a similar acute tapered rostrum butLevantocarislacks submedian gastric carinae (Galil and Clark,1993).ParaxiusaltusBate,1888,has a short toothed rostrum and almost no gastric carinae(Sakai and de Saint Laurent,1989).AustralocarispinjarupPoore &Collins,2009,lacks teeth on all gastric carinae and has a scaphocerite with ventral spines.
The arrangement of teeth on the gastric carinae ofCarolinaxiuskexuaegen.et sp.nov.resembles that of some other axiid genera such asEutrichochelesWood Mason,1876,ParaxiopsisDe Man,1905,CalaxiusSakai &de Saint Laurent,1989,andManaxiusKensley,2003,but these all have pigmented eyes on stalks and prominent supraocular spines (see Poore and Collins (2015);Poore (2020) for discussion of these genera).
LeonardsaxiusSakai,2011,andAxiopsisBorradaile,1903,have well developed eyes but otherwise resembleCarolinaxiusgen.nov.in having an acute tapering rostrum with dentition incorporating an undifferentiated supraocular spine.Species ofLeonardsaxiuslack dentition on the submedian and lateral gastric carinae.Species ofAxiopsishave rows of teeth on all gastric carinae.
About half of all axiid genera possess a male pleopod 1 as doesCarolinaxiuskexuaegen.et sp.nov.Species of the ‘calocaridid’ group have a twoarticled male pleopod 1 as does our new species but in most others,such asLitoraxiusKomai &Tachikawa,2007 andPlanaxiusKomai &Tachikawa,2008,there is a single or fused article.The second article of the male pleopod 1 inBouvieraxiusSakai &de Saint Laurent,1989 has a mesial lobe absent in the new species,while inCoralaxiusit is oval.The male pleopod 1 ofSpongiaxiusandLeonardsaxiusSakai,2011 resembles that of the new species but both genera have very different carapace sculpture.OnlyMontanaxiusmediumquodDworschak,2016 andM.vadensisPoore,2020 also have similar male pleopods 1.Both species ofMontanaxiuslack median gastric carinae and teeth on other carinae (Dworschak,2016;Poore,2020).Montaxaxiusis sistergenustoCarolinaxiusgen.nov.on the admittedly limited phylogenetic tree.
4.2 Phylogenetics
Our phylogenetic analyses are based on two mitochondrial genes (12Sand16S rRNA) and one nuclear protein coding gene (H3),which have moderate substitution rates (Toon et al.,2009) and have been suggested suitable for phylogenetic inferences at different taxonomic levels in molecular systematic studies of Axiidea (Felder and Robles,2009;Robles et al.,2009,2020;Robles and Felder,2015).
In our phylogenetic tree,the Axiidae is recovered as a well-supported monophyletic group,consistent with results of previous molecular phylogenetic studies (Tsang et al.,2008;Felder and Robles,2009;Robles et al.,2009).Carolinaxiuskexuaegen.et sp.nov.is positioned topologically as a sister group toMontanaxiusvadensisPoore,2020 in both ML and BI analyses but was weakly supported.This close relationship between the new genus andMontanaxiusDworschak,2016,is also supported by their resemblance in morphology (see above) and habitat(seamounts).Besides,Carolinaxiusgen.nov.could be associated with deep-sea hexactinellid sponges,likeMontanaxius,as the new axiid was accidentally found in the sampling basket also containing a number of deep-sea hexactinellid sponge specimens (Dr.Xuwen WU,pers.Comm.).However,the low support values (BP=67%,PP=0.67) and significant12Sand16S rRNAgenetic distances (18.9% and 20.8%,respectively) indicate they should belong to different genera.
In addition,our phylogenetic analyses revealed multiple origins of the current deep-sea axiid shrimps.Specifically,AxiopsisBorradaile,1903,a shallowwater group,is located at a basal position of the Axiidae clade.Two deep-water clades,“Carolinaxiusgen.nov.+Montanaxius”and“Acanthaxius+Bouvieraxius+Manaxius”are separated and at more derived positions in the phylogenetic tree.Our analysis is in accordance with recent findings of multiple origins of many other deep-sea crustacean taxa,such as isopods (Lins et al.,2012),mysids (Kou et al.,2020),stenopodidean shrimps (Chen et al.,2016) and caridean shrimps (Kou et al.,2016).However,many internal phylogenetic relationships within Axiidae,a family of over 200 species in almost 50 genera (WoRMS,2020),remain unresolved.Future studies with greater representation of taxa and genes will be necessary to elucidate the evolution of axiidean shrimps.
5 DATA AVAILABILITY STATEMENT
The datasets generated for this study can be found in the GenBank (http://www.ncbi.nlm.nih.gov/genbank).ZooBank registration publication LSID:urn:lsid:zoobank.org:pub:04689387-F68B-47C5-BE73-C1C4F54246FB.
6 ACKNOWLEDGMENT
We sincerely thank Dr.Kuidong XU (Institute of Oceanology,Chinese Academy of Sciences,Qingdao)for providing us with the deep-sea specimen collected during the M8 seamount cruise and Dr.Xuwen WU(Institute of Oceanology,Chinese Academy of Sciences,Qingdao) for offering us color photo and collection information of the new species.We want to express our appreciation to the crews of the R/VKexuefor their great support in sampling with ROVFaxianduring the expedition in the Caroline Ridge,Northwest Pacific in June 2019.We are also grateful to Laure Corbari,Paula Lefèvre-Martin and Anouchka Krygelmans for help in making collections and tissue samples available at Muséum National d’Histoire naturelle,Paris.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Screening of stable internal reference genes by quantitative real-time PCR in humpback grouper Cromileptes altivelis*
- Morphology and multifractal features of a guyot in specific topographic vicinity in the Caroline Ridge,West Pacific*
- Geochemical characteristics and geological implication of ferromanganese crust from CM6 Seamount of the Caroline Ridge in the Western Pacific*
- Deep-sea coral evidence for dissolved mercury evolution in the deep North Pacific Ocean over the last 700 years*
- Physical oceanography of the Caroline M4 seamount in the tropical Western Pacific Ocean in summer 2017*
- Characteristics and biogeochemical effects of oxygen minimum zones in typical seamount areas,Tropical Western Pacific*
