Combining simplified DNA extraction technology and recombinase polymerase amplification assay for rapid and equipment-free detection of citrus pathogen Phytophthora parasitica
2021-08-12CHENWeiyuYUJiaXUHengLUXinyuDAlTingtingTlANYueSHENDanyuDOUDaolong
CHEN Wei-yu ,YU Jia ,XU Heng ,LU Xin-yu ,DAl Ting-ting ,TlAN Yue-e,SHEN Dan-yuDOU Dao-long
1 Nanjing Plant Protection and Quarantine Station,Nanjing 210036,P.R.China
2 Department of Plant Pathology,Nanjing Agricultural University,Nanjing 210095,P.R.China
3 College of Chemical and Biological Engineering,Taiyuan University of Science and Technology,Taiyuan 030024,P.R.China
4 College of Forestry,Nanjing Forestry University,Nanjing 210037,P.R.China
5 Department of Plant Protection,Henan University of Science and Technology,Luoyang 471000,P.R.China
Abstract Foot and root rot caused by Phytophthora parasitica is a substantial threat to citrus cultivation,affecting both yield and quality.Thus,rapid and accurate detection of P.parasitica plays an important role in disease management.The aim of this study was to develop a simple diagnostic method to detect P.parasitica infection by combining recombinase polymerase amplification and lateral flow strips (LF-RPA).To establish the LF-RPA assay of P.parasitica,the primers and probe designed based on the Ypt1 gene were tested for specificity to P.parasitica,which showed no cross-reactivity with DNAs of other related oomycete species.The LF-RPA assay detected the amount of genomic DNA of P.parasitica which was as low as 1 pg.To make the LF-RPA assay useful in low-resource settings,four simplified DNA extraction methods were compared,after which the LF-RPA assay was applied,with no specialized equipment,to analyze a diverse range of citrus tissues by using a simplified PEG-NaOH method for DNA extraction.This method was successful in detecting P.parasitica in infected plant samples within 30 min.Combining the LF-RPA assay and a simplified DNA extraction method could be a potential detection test for P.parasitica,especially in areas with limited resources.
Keywords:citrus disease,Phytophthora parasitica,recombined polymerase amplification,equipment-free detection
1.lntroduction
Citrus is one of the most widely cultivated types of fruit worldwide,and it is also expected to be the primary fruit industry in China.According to the statistics of the Ministry of Agriculture and Rural Affairs of the People's Republic of China,both the planted area and yield of citrus increased in 2017 compared to 2016,reaching 2.58 million ha and 38.16 million tons,respectively.Notably,it has been predicted that the planted area and yield of citrus in China will increase stably.However,several diseases caused by various plant pathogens have resulted in significant annual economic losses and also threatened food security globally (Dalioet al.2017).Phytophthoraspp.cause severe and economically important soil-borne diseases of citrus crops in China and worldwide (Jagtapet al.2012;Puglisiet al.2017;Yanet al.2017).Foot and root rot occurs when an infection near the ground leads to bark lesion extension.Root rot results from infection of the cortex of fibrous roots,which turns soft and becomes water-soaked.It has been reported that more than 10Phytophthoraspecies can infect citrus,butP.parasiticais considered the most destructive due to its wide distribution and broad host range (Panabiereset al.2016).
Phytophthora parasiticais a hemibiotrophic pathogen that is widespread worldwide (Erwin and Ribeiro 1996).It is capable of infecting a broad range of plants,ranging from crop plants to fruit trees (Clineet al.2008;Kamounet al.2015;Panabiereset al.2016).Phytophthora parasiticais especially aggressive toward citrus,leading to foot and root rot and causing severe damage to the trees.Phytophthora parasiticainitially infects the plants as a biotrophic pathogen and then colonizes by necrotrophic agent to disseminate from necrotic tissues (Glazebrook 2005).Infection of the main branches of citrus plants results in damage to the cambium and inner bark and the formation of cankers with gum exudation (Alvarezet al.2009;Graham and Feichtenberger 2015).Infected plants lack vigor,have reduced yield,and may even die (Queiroz and Melo 2006).Phytophthora parasiticaproduces both asexual zoospores and sexual oospores (Menget al.2014).The oospores and thick-walled resting chlamydospores are conducive to long-term survival under disadvantageous conditions and are the primary source of infection in a new season(Menget al.2014).The zoospores contribute to longdistance dissemination,which leads to repeated infection of hosts and rapid spread in fields (Menget al.2014).In addition,P.parasiticaadapts well to climate warming,and the significance ofP.parasiticais increasing under global climate warming (Kamounet al.2015;Panabiereset al.2016).
Currently,integration of cultural practices and methods of chemical control are necessary to managePhytophthorainduced foot and root rot.The cultural practices include the application of tolerant rootstocks and proper irrigation practices.Before adopting a chemical control method,sensitive and accurate detection should be a prerequisite for managing the disease caused byP.parasitica.Molecularbiology-based detection technologies provide effective and timesaving methods for detecting plant pathogens.Diverse molecular detection methods have been established to detectP.parasitica,including nested PCR,multiplex PCR,and real-time quantitative PCR (Ippolitoet al.2002,2004;Daset al.2013;Blayaet al.2015).Detection ofP.parasiticafrom various samples (irrigation water,soil and plant tissues) has been achieved using these methods(Konget al.2003;Meng and Wang 2010).Further,detection methods based on the isothermal amplification technique have developed rapidly in recent years.Liet al.(2015)set up a loop-mediated isothermal amplification (LAMP)assay ofP.parasiticaand applied it effectively.Compared to the LAMP assay,recombinase polymerase amplification(RPA) technology has more advantages,including fewer primers (two primers),a faster amplification time (15 min),and a lower incubation temperature (37–42°C) (Lobato and O’Sullivan 2018).RPA is one of the most promising methods that can be applied in the field (Donoso and Valenzuela 2018).Recent studies have indicated that the Ras-related protein (Ypt1) gene is an effective target for detection ofP.parasitica(Meng and Wang 2010;Liet al.2015) and other pathogens belonging to the genusPhytophthora(Zhaoet al.2015;Khanet al.2017).
The need for in-field diagnosis of plant diseases by nonprofessional farmers has been increasing,and the RPA reaction provides a simple method for amplifying target sequences.However,to make this promise a reality,simplified sample preparation and reading of results are still required.Considerable efforts have been made to produce an equipment-free detection procedure by using simplified nucleic acid extraction.The alkaline polyethylene glycol(PEG-NaOH) extraction method has been used widely in crude extraction of nucleic acids (Hodgettset al.2011;Silvaet al.2018),and another nucleic acid extraction technology based on untreated cellulose-based paper (Whatman 1)also has promise for applications (Zouet al.2017).Both nucleic acid extraction methods require no centrifugation,so they are not limited by specialized equipment or a need for electricity.Furthermore,assessment of detection results by unaided eye is the most-effective and feasible method.SYBR Green I,an asymmetrical cyanine dye and the triggered chromism reaction which can visualize amplicons,have been used commonly to read the visual results (Caoet al.2018).Moreover,lateral flow strips have typically been used to indicate the reaction results (Yinet al.2017;Solimanet al.2018).This detection method will become much more available when simplified sample preparation,RPA amplification,and results reading are combined.
This study developed an equipment-free visual detection method forP.parasiticaby targeting theYpt1gene.This detection method was established based on the LF-RPA assay by combining simplified DNA extraction methods.Specificity and sensitivity assays were conducted after optimization of the different reaction parameters.Moreover,infected plant samples were used to evaluate the effectiveness of the LF-RPA assay.
2.Materials and methods
2.1.Microorganism culture and DNA extraction
This study used:three isolates fromP.parasitica(clade 1);21 isolates from 11 otherPhytophthoraspecies,includingP.mirabilis(clade 1),P.ipomoeae(clade 1),P.cactorum(clade 1),P.infestans(clade 1),P.capsici(clade 2),P.cryptogea(clade 8),P.palmivora(clade 4),P.sojae(clade 7),P.drechsleri(clade 8),P.litchi(clade 4),andP.megasperma(clade 6);and 26 isolates from 15Pythiumspecies,includingPy.spinosum,Py.aphanidermatum,Py.helicoides,Py.ultimum,Py.irregulare,Py.arrhenomanes,Py.hydnosporum,Py.dissotocum,Py.catenulatum,Py.intermedium,Py.marsupium,Py.splendens,Py.heterothallicum,Py.sylvaticum,andPy.oligandrum.Oomycete species were cultured individually in 10% V8 medium in the dark.Unless otherwise indicated,DNA was extracted from the mycelia.DNA extraction was accomplished by following the manufacturer’s instructions supplied in the E.Z.N.A.®HP Fungal DNA Kit (Omega Biotek,Omega Biotek,Norcross,USA),and the materials were then stored at–80°C.Genomic DNA was tested using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific,Thermo Scientific,USA).
2.2.Design of primers and probe for LF-RPA
TheYpt1gene was selected as the target gene for designing the primers and probe for the LF-RPA assay ofP.parasitica.TheYpt1gene sequences of diversePhytophthoraspecies were downloaded from the GenBank Database,includingP.parasitica(GenBank accession numbers:KR632915,MK058408,MH988452,MH988453,and JN678924),P.cactorum(MG880404),P.ipomoeae(KR632905),P.infestans(KR632902),P.hedraiandra(MH443241),P.idaei(JN678922),P.pseudotsugae(JN678925),P.clandestine(JN678920),P.iranica(JN678923),P.tentaculata(JN678926),P.mirabilis(KR632910),P.phaseoli(KR632911),P.capsici(MH975004),P.sojae(MG880402),P.palmivora(MK058414),P.cryptogea(KJ755146),P.megasperma(KJ755178),P.katsurae(DQ162980),P.pseudosyringae(DQ162967),P.polonica(MK058420),andP.kernoviae(DQ270322).Then,multiple alignment ofYpt1gene sequences was performed.The unlabeled forward primer PpRPALF-F(5´-CTATTCAACTAACCGATGTACCAAATCACGTGTGT-3´)and the biotin-labeled reverse primer PpRPALF-R (5´-Biotin-GCTCTTTTCCTTGGATCTTCTCTCGATAAGTCGG-3´)were designed according to the specific region ofP.parasitica(Appendix A).For the specific probe PpRPALF-P (5´-FAMCGACGTGACGGACCAGGAGTCATTCAACAA-THFGTGAAACAGTGGCTG-C3 space-3´),the 5´ end was FAM labeled and the 3´ end was blocked with a phosphate group,which was inserted between the 30th and 31st base.Then,the specificities of the primers,PpRPALF-F and PpRPALF-R,and probe were confirmed further by the NR Database of the NCBI.The specific primers and probe PpRPALF-P were synthesized by Sangon Biotech(Shanghai,China).
2.3.Establishment and optimization of the LF-RPA
One kind of commercial lateral flow (LF) strip,GenLine HybriDetect MGHD1,supplied by Milenia Biotec (Germany),was applied to detect RPA amplicons visually.The LF strips were designed to detect amplicons labeled with biotin and 6-carboxy-fluorescein (FAM),which were obtained by using a TwistAmp nfo Kit (TwistDX,UK).The LF-RPA reaction mixture had a total volume of 50 μL which contained 2 μL of DNA template,2.1 μL of each primer (10 μmol L−1),0.6 μL of probe PpRPALF-P (10 μmol L−1),11.2 μL of ddH2O,29.5 μL of rehydration buffer,and 2.5 μL of 280 mmol L−1MgSO4.The MgSO4was initially added to the cap of the tube containing the freeze-dried pellet supplied in the TwistAmp nfo Kit.The tubes were incubated at 40°C for 20 min after brief vortexing and short centrifugation.Reaction products (5 μL) were added directly on the sample pad of the strips,and then the strips were vertically immersed into separate centrifuge tubes containing 100 μL of HybriDetect Assay Buffer.The results of the RPA reaction could be determined by unaided eye after 5 min.The C band (control band) must appear to ensure that strips are usable,and the T band (test band)represents a positive result.
The reaction conditions for the LF-RPA assay were optimized.First,different groups of reverse primer PpRPALF-R (10,5,and 2.5 μmol L−1) and probe PpRPALF-P(10 and 5 μmol L−1) concentrations were evaluated (Table 1).Second,different loading volumes of amplification products(1,2,and 5 μL) added to the strips were evaluated.Moreover,various incubation temperatures (25,30,35,40,45,and 50°C) and incubation times (5,10,20,30,40,and 50 min) for the LF-RPA were tested.
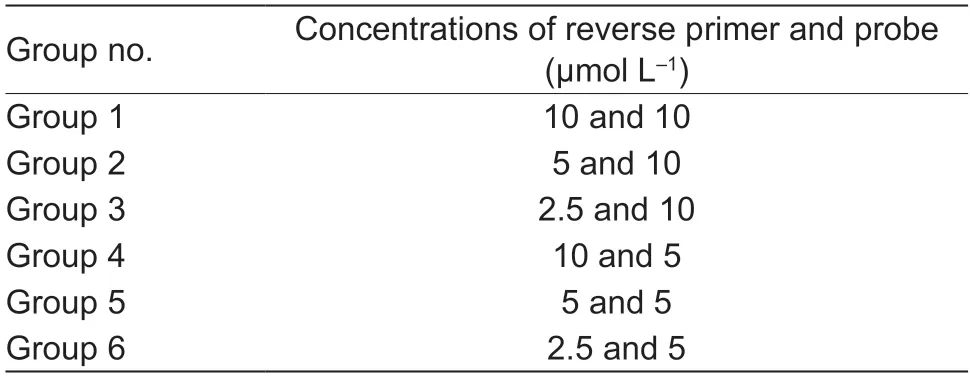
Table 1 Comparison of various combinations of reverse primer and probe concentrations
2.4.Specificity and sensitivity of LF-RPA
The specificities of the primers PpRPALF-F/PpRPALF-R were tested using three isolates fromP.parasitica,21 isolates from 11 otherPhytophthoraspecies,and 26 isolates from 15Pythiumspecies.All genomic DNA of the isolates was guaranteed by using primers ITS1 and ITS4 (Whiteet al.1990) in conventional PCR,which wascarried out byEx Taqpolymerase (TaKaRa Bio.,Japan),as described by the supplier.For the sensitivity assay,a 10-fold serial dilution ofP.parasiticaDNA ranging from 10 ng to 10 fg was amplified in the RPA reaction by using the TwistAmp Basic Kit (TwistDX,UK).Double-distilled water(ddH2O) was used as the negative control.The RPA assay was carried out under optimal conditions.
2.5.Simplified DNA extraction methods
An equal amount of mycelia tissue was extracted individually by four simplified DNA extraction methods.First,the mycelia tissue was ground for 1 min in a 1.5-mL tube with lysis buffer,which contained 900 μL 2% CTAB,90 μL 10% SDS,and 10 μL β-mercaptoethanol and the following procedure was conducted as described in a previous study (Doyle and Doyle 1990).The DNA obtained was finally dissolved in 500 μL ddH2O.Second,the DNA was directly extracted by ddH2O,as described in a previous study (Takahashiet al.2014).The details follow.Sample tissues (50 μL)were placed into a 1.5-mL reaction tube containing 450 μL ddH2O,which was then shaken by hand for 1 min and incubated for 2 min.Third,PEG-NaOH was used as the lysis buffer to extract DNA,as described by Silvaet al.(2018).The sample (50 μL) was immersed in a tube in 450 μL of alkaline-PEG buffer (6% PEG 200 with 20 mmol L−1NaOH),which was freshly prepared.The tubes were shaken by hand for 1 min and incubated for 2 min.Fourth,the extraction method was applied following the protocols proposed by Zouet al.(2017).Tissue (50 μL) was added to a tube in the presence of 450 μL of extraction buffer #1(Zouet al.2017) and incubated for~30 s.
For a comparison of the four methods,2 μL of extraction solution was diluted in 198 μL of extraction buffer #1.A 3-mm diameter disc of Whatman 1 filter paper was placed into the extraction tube for 30 s.It was then transferred to 200 μL of wash buffer (Zouet al.2017) for 1 min by pipette tip.To estimate the quantity and quality of DNA extracted by the four methods mentioned above,5 μL of the extraction solution was first analyzed by electrophoresis in 1%agarose gels.For a better comparison of the amplification efficiency of the extracted DNA,a serial 10-fold dilution of the extraction solution was further assessed by RPA reaction.Solution (2 μL) was amplified separately using the TwistAmp Basic Kit.For the fourth method,using Whatman 1 paper,the disc-captured DNA from each dilution was carried directly into an amplification reaction tube.
To evaluate the DNA extraction methods using PEG-NaOH and Whatman 1 paper further,leaves of 3-weekNicotiana benthamianawere inoculated with freshP.parasiticamycelia.The infected leaves were photographed under ultraviolet light after 36 h of infection and then divided into four sections (I,II,III,and IV).The same area of each section was collected for DNA extraction by the two methods with 200 μL of extraction buffer.Finally,2 μL of the DNA extracted by using the PEG-NaOH and Whatman 1 disc was amplified as the template in LF-RPA.
2.6.Equipment-free detection of infected samples
Phytophthora parasiticainoculations of citrus leaves,fruits,and fresh branches were conducted as described in a previous study (Yanet al.2017).The leaves,fruits,and branches were cleaned with sterilized water,dipped into 70% ethanol for~10 s,and then washed with sterilized water (Tianet al.2019a,b).The surface-sterilized citrus leaves and fruits were then stabbed once with a sterilized needle.Furthermore,a small cut of 1 mm×1 mm extending to the xylem tissue was made in the citrus branches.FreshP.parasiticaculture was placed on the lesion sites of leaves,fruits,and branches.The inoculated samples were then placed on filter papers drenched with sterilized water to maintain humidity and kept in the dark at 25°C.The extent of symptoms of infection was recorded after inoculation.The samples were divided into three sections (I,II,and III) and analyzed by LF-RPA assay using the PEG-NaOH extraction method.
To evaluate the LF-RPA assay further,suspectedP.parasitica-infected tissue samples from citrus main branches were collected during March in Jiangxi Province,China.Then,the samples were analyzed using the LF-RPA assay combined with PEG-NaOH extraction method.
3.Results
3.1.Optimal reaction conditions of LF-RPA
To obtain optimal reaction conditions for the LF-RPA assay,several parameters of LF-RPA,including proportions of primer PpRPALF-R and probe PpRPALF-P,incubation temperature,and loading volume of reaction products on LF strips,were evaluated.To ensure maximization of primer and probe efficiencies,first,different combinations of proportions of reverse primer PpRPALF-R and probe PpRPALF-P were analyzed (Table 1).As shown in Fig.1-A,test bands were hardly observed for groups 3,4,5,and 6,which indicated low amplification efficiency.However,clear bands were obtained for the remaining two groups.Notably,a bright test band in comparison to other groups was produced by group 1.Hence,the highest efficiency combination of reverse primer PpRPALF-R and probe PpRPALF-P was 10 μmol L−1for both.Second,the loading volumes of reaction products (1,2,and 5 μL) were assessed.The loading volumes of 2 and 5 μL produced test bands on LF strips,but the band was much clearer when a volume of 5 μL was used (Fig.1-B).Thus,5 μL of amplicon was added to the LF strip in further analyses.
The RPA reaction is commonly conducted at 37–42°C.However,it is thought that amplification could occur at room temperature without any extra heating device or by using body warmth.To evaluate the temperature range of the LFRPA reaction,amplification was conducted under a serial temperature gradient from 25 to 50°C.Under the lowest(25°C) and highest (50°C) temperatures,the RPA reaction failed to produce amplicons.However,at 30,35,40,and 45°C,distinct test bands were produced by the LF-RPA assay (Fig.1-C),which indicated that the LF-RPA method is highly efficient within a broad range of constant reaction temperatures between 30 and 45°C.However,the method seemed to be the best at 35–40°C (Fig.1-C).Considering the variability of field temperatures,it is necessary to guarantee a suitable temperature for the LF-RPA reaction.
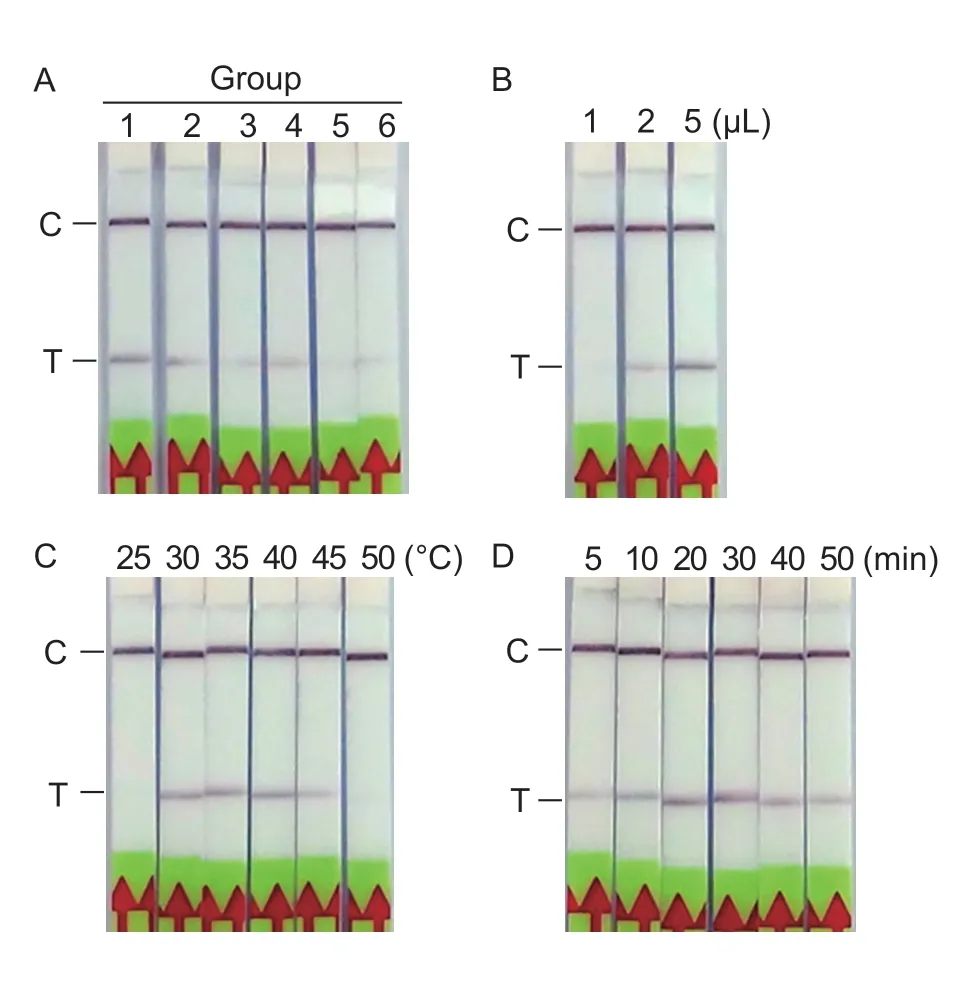
Fig.1 Optimization of the LF-RPA assay.A,optimization of reverse primer and probe concentrations.Six combinations containing different ratios and concentrations of reverse primer and probe were evaluated in the LF-RPA assay.The detailed information for each group is summarized in Table 1.B,optimization of the volume of RPA amplicons added onto LF strips.C,optimization of the RPA incubation temperature.D,amplification time of the LF-RPA assay.LF-RPA amplification was carried out at 40°C for different time periods,followed by incubation of the LF strips at room temperature for 5 min.C,control band;T,test band.
To evaluate LF-RPA amplification time,10 ng of genomic DNA ofP.parasiticawas amplified at 40°C for various time periods (5,10,20,30,40,and 50 min),followed by incubation of LF strips at room temperature for 5 min.The test band was observed after a 10-min amplification time(Fig.1-D).With prolongation of amplification time,the amplicons accumulated,making the test band clearer.However,the amplicons that could be detected by LF strips were reduced after 50 min of amplification,as shown by the test band.Considering the detection efficiency,a 20–30 min amplification time should be suitable forP.parasiticadetection.
3.2.Detection specificity of LF-RPA
The specificities of the primers and probe designed forP.parasiticawere first confirmed by performing a Blast search against the NCBI NR database.To access the detection specificity further,the DNA extracted from the three isolates fromP.parasitica,21 isolates from 11 otherPhytophthoraspecies,and 26 isolates from 15Pythiumspecies were amplified individually as templates in RPA reaction (Fig.2).Amplification bands were observed only for the three isolates fromP.parasitica.No bands were obtained from the other testedPhytophthoraorPythiumisolates.The results indicated that the designed primers PpRPALF-F,PpRPALF-R and probe PpRPALF-P were highly specific forP.parasiticaand can be applied to detect differentP.parasiticaisolates.
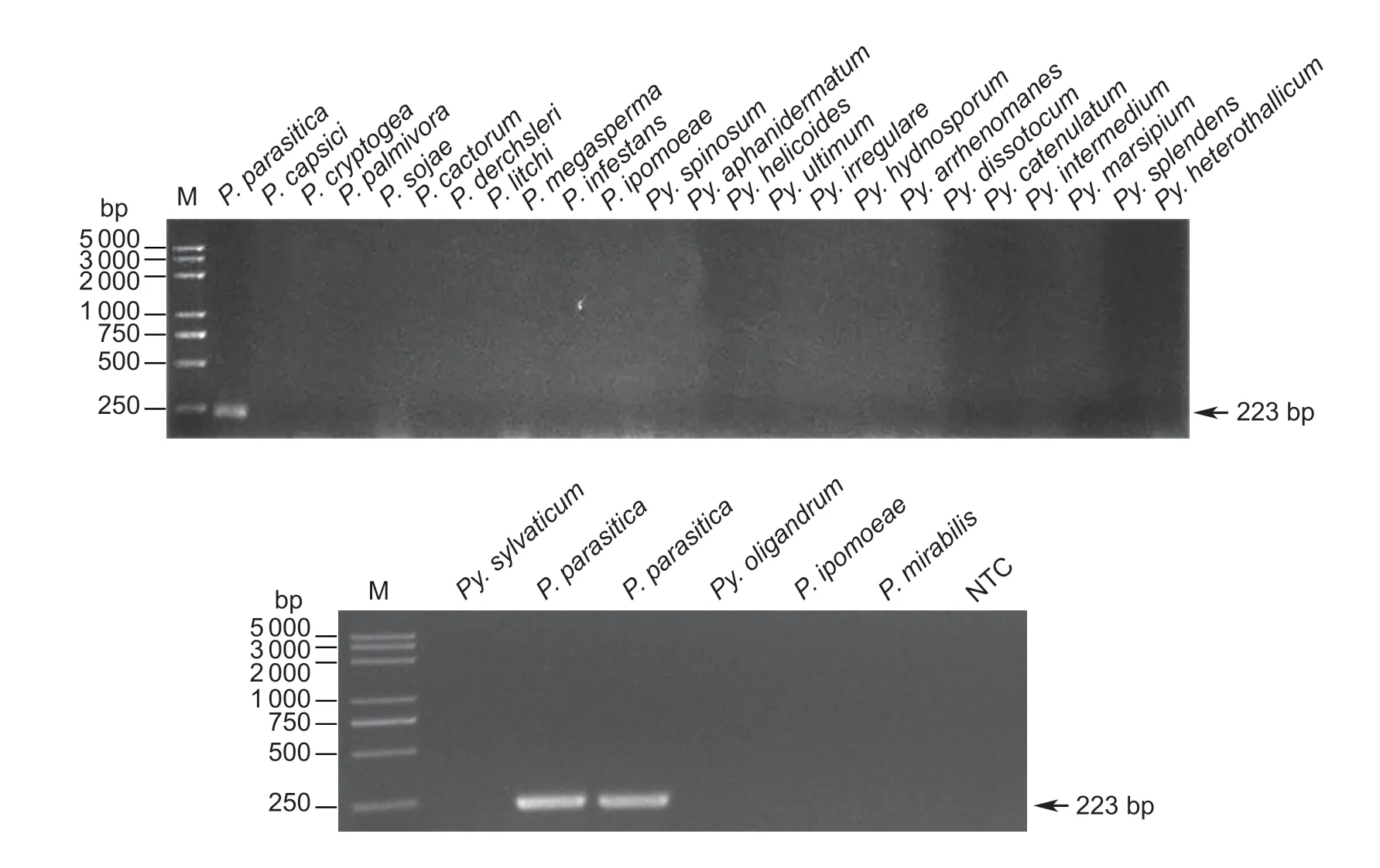
Fig.2 Specificity of the LF-RPA assay.RPA was conducted with the primer pair PpRPALF-F/R.In total,three isolates from Phytophthora parasitica,11 other Phytophthora species,and 15 Pythium species were used in the specificity assay.Lane M,100-bp DNA ladder;lane NTC,negative control.
3.3.Detection sensitivity of LF-RPA
In order to evaluate the sensitivity of the LF-RPA reaction,serial dilutions ofP.parasiticagenomic DNA,varying from 10 ng to 10 fg,were tested.The same volume of sterilized water was used as the negative control (NTC).At least three repetition tests were carried out.As shown in Fig.3,the sensitivity of LF-RPA was as low as 1 pg of genomic DNA,whereas the detection sensitivity of basic RPA was 100 pg.The results indicated that LF-RPA,by combination with LF strips,is more sensitive than basic RPA.
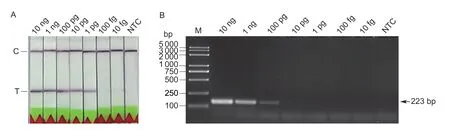
Fig.3 Detection sensitivity of LF-RPA assay. Various amounts of genomic DNA of Phytophthora parasitica were applied to evaluate the detection limits of the LF-RPA assay (A) and the basic RPA assay (B).Lane M,2K plus DNA ladder;lane NTC,negative control;C,control band;T,test band.
3.4.Evaluation of simplified DNA extraction methods
To select the most suitable DNA extraction technique for LF-RPA outside the laboratory,such as on farmland,four different methods performed using CTAB,H2O,PEG-NaOH and Whatman 1 paper were used to extract genomic DNA fromP.parasiticamycelia.First,extracted DNA was estimated by agarose gel electrophoresis.However,the DNA captured by the Whatman 1 disc could not be analyzed by this method.Less DNA was extracted by H2O than by the other two methods (Fig.4-A).Similar amounts of DNA were obtained by the PEG-NaOH and CTAB methods,but the DNA quality was higher in the former.The CTAB method is a conventional method for extracting DNA;however,it requires centrifugation and other organic reagents.The PEG-NaOH method is a much faster extraction procedure.Second,the amplification efficiency of the RPA reaction was evaluated using a 10-fold dilution of the DNA extracted by the four methods.As shown in Fig.4-B,the PEG-NaOH method had the highest amplification efficiency.This result is consistent with the DNA amounts and quality shown in Fig.4-A.In conclusion,the PEG-NaOH method is a simple and timesaving method for extracting DNA,and it has promise for use in the field.
In a natural setting,it is more likely to detectP.parasiticafrom infected plant samples.Nevertheless,the tests of the four methods may have undervalued the extraction efficiency of the Whatman 1 method.To facilitate comparison,the 3-mm Whatman 1 disc was added into the extraction solution to capture DNA.The total amount of DNA in the extraction solution was equal to 2 μL/500 μL of the extraction solution amount of the other three methods.As for the efficiency of DNA capture and release,the amplification template DNA method certainly had a lower result than the other three methods.Furthermore,the DNA contained in the extraction buffer was much higher than what was set in most cases.Hence,artificially inoculated plant samples were used to evaluate the PEG-NaOH and Whatman 1 methods further.The infected leaves were divided into four regions (regions I to IV),with region I being closest to the inoculation site(Fig.4-C).The LF-RPA assay detectedP.parasiticawhen no symptoms were observed by unaided eye (regions II and III),indicating high detection sensitivity.Moreover,there were few differences between the two methods.Thus,neither the PEG-NaOH nor Whatman 1 method alone is suitable for use in equipment-free detection.
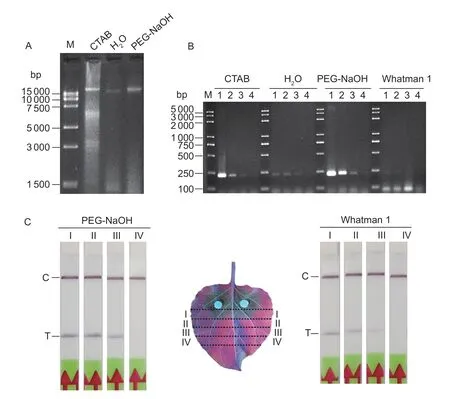
Fig.4 Comparison of DNA extraction methods.A,genomic DNA obtained by the CTAB,H2O and PEG-NaOH methods was analyzed by agarose gel electrophoresis.B,serial dilutions of the extraction solutions obtained by the CTAB,H2O,PEG-NaOH,and Whatman 1 methods were used as a template in basic RPA amplification.Lane 1,original extraction solution;lanes 2–4,10,100,1 000 times dilutions of the original extraction solution.C,LF-RPA was conducted to analyze infected samples using the PEG-NaOH and Whatman 1 methods.The infected leaves were divided into four sections (I,II,III,and IV) for DNA extraction and the LF-RPA assay.C,control band;T,test band.
3.5.Equipment-free detection of infected samples
To evaluate the LF-RPA assay further,first,different tissues of artificially inoculated citrus were assessed by LF-RPA.As shown in Fig.5-A,B,and C,P.parasiticaDNA was amplified and detected visually in infected areas (region I)of different tissues (leaves,branches,and fruits) by the PEG-NaOH extraction method.However,only fruit without symptoms was detected positively (Fig.5-C,region II).This may have been related to the infection speed ofP.parasiticain different tissues,because it was found that symptoms appeared on citrus fruits at 2 days after inoculation but on leaves or branches at 5–7 days after inoculation.Second,suspectedP.parasitica-infected tissue samples from the main branches of citrus were detected by the LF-RPA assay.The samples were gathered in March from a citrus orchard in Jiangxi Province,China,which is one of the citrus producing areas in the country.The temperature after prolonged rainfall was between 7 and 18°C.Among the 10 samples collected,three produced positive results using both of LR-RPA and conventional PCR methods (Fig.5-D).
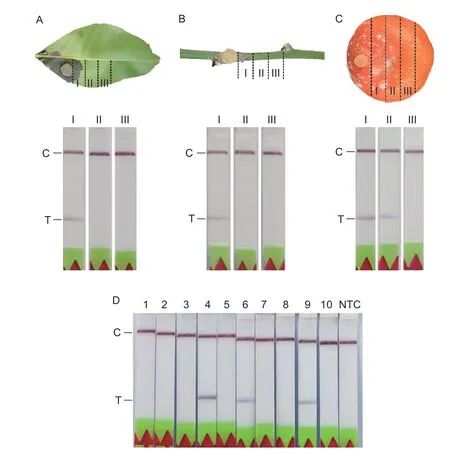
Fig.5 Equipment-free detection of infected samples using LF-RPA assay for Phytophthora parasitica.Infected citrus leaf (A),branch (B) and fruit (C) tissues were divided into three sections (I,II,and III) and detected individually.D,10 suspected infestation tissue samples from citrus main branches were assessed.C,control band;T,test band.
4.Discussion
Due to the importance of the citrus industry in China and in the world,diseases caused by plant pathogens have attracted progressively more attention.Citrus Huanglongbing (HLB)is one of the most destructive diseases,and it is spreading rapidly in citrus-producing areas worldwide.HLB pathogens infect all parts of citrus tree including the roots (Tatineniet al.2008).Studies have indicated that the increasing incidence of HLB in the presence ofP.parasiticamay result in a greater impact on fibrous root health than the HLB pathogen (Las) alone (Wuet al.2017).Therefore,effective management ofP.parasiticamay also benefit the control of HLB in citrus orchards.Rapid and accurate detection ofP.parasiticawould play an important role in this effort,and different molecular detection technologies based on PCR forP.parasiticahave been developed and used widely.The detection limit of nested PCR assays through two rounds of amplification is up to 100 fg when targeting theYpt1gene (Liet al.2015).Quantification assays ofP.parasiticahave been realized by using real-time PCR with the TaqMan probe (Blayaet al.2015).However,these diagnostic methods based on PCR require specialized instruments and professional staff in a laboratory.These technologies cannot be applied outside the laboratory in resource-limited settings.However,the development of isothermal amplification techniques in recent years has been a crucial advance in field-based plant-disease diagnosis.In 2015,the LAMP assay ofP.parasiticawas developed (Liet al.2015).Compared to the LAMP assay,the established LF-RPA assay forP.parasiticarequires less time,fewer primers,and lower incubation temperatures.
Some kinds of citrus trees can be cultivated in a greenhouse,but the high temperature and humidity may lead to heavier infection byP.parasitica.Thus,accurate and rapid diagnosis of the diseases caused byP.parasiticainfection plays a crucial role in disease management.The RPA assay is a promising method for use in field-based molecular diagnosis.However,DNA extraction without special instruments is a key issue,so this problem must be addressed before field-based application is possible(Donoso and Valenzuela 2018).Attempts were made to simplify the nucleic acid purification progress,and the PEGNaOH-based DNA extraction method was subsequently modified and used in various DNA-based tests (Hodgettset al.2011;Silvaet al.2018).Zouet al.(2017) developed a simple extraction approach by using a cellulose-based dipstick and obtained nucleic acid in 30 s.Considering the good compatibility of samples with the RPA reaction,this study applied and compared four different DNA extraction methods.The results indicated that the PEG-NaOH method and cellulose-based dipstick (Whatman 1) method could both be carried out to prepare crude DNA for the LF-RPA assay.Furthermore,the LF-RPA assay effectively detectedP.parasiticafrom artificially infected (leaves,branches,and fruits) and naturally infected citrus tissues.
5.Conclusion
In this study,the LF-RPA assay was developed for rapid and accurate detection ofP.parasiticawith a high level of specificity and sensitivity.In combination with a simplified DNA extraction method,the entire detection could be completed in 30 min without any specialized equipment,and it was applied successfully for artificially and naturally infected plant samples.Thus,the LF-RPA assay is a promising method forP.parasiticadetection in the field and in resource-limited areas.
Acknowledgements
We are grateful to Dr.Dong Suomeng,Department of Plant Pathology,Nanjing Agricultural University,China,for providingPhytophthora mirabilisisolate.This work was funded by grants from the Fundamental Research Funds for the Central Universities,China (KYT202001 and JCQY201901) and the Special Fund for Agro-scientific Research in the Public Interest,China (201503112).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendixassociated with this paper is available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Assessing the impact of non-governmental organization’s extension programs on sustainable cocoa production and household income in Ghana
- Triple bottom-line consideration of sustainable plant disease management:From economic,sociological and ecological perspectives
- A rice geranylgeranyl reductase is essential for chloroplast development
- lmpacts of climate change on drought risk of winter wheat in the North China Plain
- Rapid determination of leaf water content for monitoring waterlogging in winter wheat based on hyperspectral parameters
- Does nitrogen application rate affect the moisture content of corn grains?
