Equilibrium folding and unfolding dynamics to reveal detailed free energy landscape of src SH3 protein by magnetic tweezers*
2021-07-30HuanhuanSu苏环环HaoSun孙皓HaiyanHong洪海燕ZilongGuo郭子龙PingYu余平andHuChen陈虎
Huanhuan Su(苏环环) Hao Sun(孙皓) Haiyan Hong(洪海燕)Zilong Guo(郭子龙) Ping Yu(余平) and Hu Chen(陈虎)
1Institute for Biomimetics and Soft Matter,Fujian Provincial Key Laboratory for Soft Functional Materials Research,Department of Physics,Xiamen University,Xiamen 361005,China
2Center of Biomedical Physics,Wenzhou Institute,University of Chinese Academy of Sciences,Wenzhou 325000,China
3Oujiang Laboratory,Wenzhou 325000,China
Keywords: protein folding and unfolding,magnetic tweezers,free energy landscape,transition state
1. Introduction
Most proteins fold to their specific native structures in physiological environment to perform their biological functions,[1]and unfold before degradation[2]or during translocation process.[3]Many diseases are caused by the misfolding or failure of degradation of damaged proteins,such as madcow disease and Alzheimer’s disease.[4,5]Therefore, revealing the basic mechanism of protein folding and unfolding is critical to development of new treatment strategy of this kind of diseases.
Amino acids sequence of protein determines its native structure as the global minimal point in its free energy landscape,[1]while the topological arrangement ofα-helix andβ-strand in the native structure and contact number of the native structure regulate the protein’s folding mechanism and folding rate.[6]Biochemical bulk experiment has been used to study protein stability and folding dynamics.[7,8]Denaturant or temperature is changed suddenly while fluorescence,circular dichroism, or nuclear magnetic resonance hydrogen exchange signals are recorded.[9]In bulk experiment,average properties of all protein molecules are measured,which makes it difficult to detect transient intermediate state, and the unfolding rate under physiological condition and free energy can only be estimated from extrapolation.
Single-molecule force spectroscopy technique has been used to study the folding and unfolding dynamics of proteins.[10-13]Molecular dynamic simulation can also be used to construct the free energy landscape of small proteins.[14]In single molecule manipulation experiment,stretching force is applied between two specific amino acids,usually the N-terminus and C-terminus of the protein. Extension is measured with nanometer resolution to monitor the state of protein. Atomic force microscopy (AFM) has been widely used to study force response of proteins at high-force regime.[15]Optical tweezers can apply low force or low loading rate to stretch proteins, and record unfolding and folding processes close to equilibrium transitions.[16]
The most stable and robust single molecule technique,magnetic tweezers, can apply intrinsic constant force from zero to more than 100 pN over hours or even longer time scale.[17-20]The critical force is defined as the force at which protein has equal folding rate and unfolding rate. Equilibrium folding and unfolding process at the neighborhood of critical force can be directly recorded, even for very stable proteins with extremely slow unfolding rate at low forces. Critical force for single domain proteins with around 50 to 150 amino acids is usually only 4-8 pN.[17,21]Therefore, such an equilibrium measurement is a good mirror of the folding and unfolding dynamics in the absence of force.
Simple two-state folding proteins have only two dominant kinds of conformations classified as the native state and unfolded state.[22]Src SH3 domain (native structure and amino acid sequence are shown in Fig. 1(a)) is a typical two-state protein of 64 amino acids including 56 amino acids forming a stable compact structure,which has been used as a model protein to study protein folding dynamics.[7,22]Native structure of src SH3 protein is composed of two three-strandedβ-sheets packed orthogonally to form a smallβ-barrel structure.[23-25]
Force spectroscopy experiment using optical tweezers found that the mechanical resistance of src SH3 to stretching force is dependent on different pulling axes: shearing pulling geometry and unzipping pulling geometry.[8,26]To design and construct hydrogel, direct pulling from N- and C-termini of src SH3 has been carried out by AFM with constant pulling speed.[27]In the optical tweezers experiment, the force range of unfolding is from 7 to about 40 pN,while the folding force range is from 4 to 7 pN,including both shearing and unzipping pulling directions.[28]However, equilibrium folding and unfolding dynamics was not reported. Theoretically the folding rate and unfolding rate in the absence of force should be independent of pulling geometry. The reported force-dependent unfolding rates along shearing and unzipping pulling axes cannot be extrapolated to the same value at zero force,which indicates that unfolding transition at lower forces may show behavior deviating from linear extrapolation.
In this paper, we report the first equilibrium folding and unfolding dynamics measurement of SH3 protein under constant forces, from which the folding free energy of SH3 is determined directly. Additionally, force-dependent folding rate,unfolding rate,and transition step size are obtained from both equilibrium measurement and force-jump measurement.Based on experimental results, a two-state free energy landscape with N-C distance as reaction coordinate is constructed with detailed parameters of folding free energy,barrier height and location.
2. Materials and methods
2.1. Sample preparation
The src SH3 (PBD: 1SRL) gene was synthesized (Gen-Script Biotech)and cloned into the vector pET151-I27 which has two Titin I27 domains on each side of the multiple cloning site.[7]Plasmids pET151 harboring His6-AviTag-I272-src SH3-I272-SpyTag and pBirA(Biotin ligase plasmid)were transformed into the E.coli strain BL21 (DE3). Transformed E.coli cells were cultivated in LB medium (supplemented with chloramphenicol, ampicillin, and D-biotin) at 37°C until the optical density (OD) of the bacterial cell reached 0.6-0.8. After applying the inducer of isopropyl-β-D-thiogalactopyranoside (IPTG) for 12 h at 25°C, the cells were harvested by centrifugation and lysed by sonication in a buffer(50 mM Tris,500 mM NaCl,50%glycerol,5 mM imidazole, 5 mM 2-mercaptoethanol, pH 8.0). The protein src SH3 was purified with Ni-NTA Sefinose(TM)Resin(Sangon Biotech)and Superdex 200(GE Healthcare),according to the manufactures’protocol,then quickly frozen in liquid nitrogen and stored at-80°C.[21]
2.2. Magnetic tweezers measurement
Coverslips were cleaned firstly by sonicator and plasma cleaner, then were silanized by methanol solution of 1% 3-aminopropyltriethoxysilane(APTES,cat. A3648, Sigma)for 1 h. Flow chambers were made by sandwiching a piece of functionalized coverslip and another piece of coverslip with parafilm in between. Polybead amino microspheres (cat.17145, Polysciences) with diameter of 3.0 μm were flowed into chamber and incubated for 20 min to get stuck on the coverslip that is used to eliminate spatial drift during the single molecule experiment. The flow chamber was filled by 1%Sulfo-SMCC(SE 247420,Thermo Science)and incubated for about 20 min, then rinsed by 200 μL PBS buffer. After that,SpyCatcher protein in PBS was flowed into the chamber and incubated for 2 h. In order to passivate the surface, 1% BSA in tris buffer pH 7.4 was flowed into chamber and incubated overnight at room temperature. Before single molecule experiment,chambers were incubated in PBS with around 1 nM protein src SH3 for 15 min. Streptavidin-coated paramagnetic beads Dynabead M270 (cat. 65305, Invitrogen) were flowed into the chamber to form protein tethers. Finally,1%BSA solution with 5 mM L-Ascorbic Acid Sodium Salt was flowed into chamber to wash out untethered beads.[29]
Home-made magnetic tweezers were used to apply stretching force to src SH3 protein tether to study its forcedependent folding and unfolding dynamics. Constant force equilibrium measurements and force-jump experiments were performed in force ranges of 3.5-6 pN and 4-11 pN, respectively. Details of magnetic tweezers design can be found in our previous publications.[10,17,21]
3. Result
3.1. Constant loading rate experiment to identify correct tether
In magnetic tweezers experiments, the recombinant protein construct of AviTag(biotin)-I272-src SH3-I272-SpyTag was linked between SpyCatcher-coated coverslip and streptavidin-coated paramagnetic bead (Fig. 1(b)). The correct src SH3 protein tether was initially verified by force-ramp experiments at constant loading rate of 0.5 pN/s. Two kinds of unfolding events were observed: the unfolding step of src SH3 protein at~5 pN and four typical unfolding steps of titin I27 with size>20 nm at forces greater than 60 pN.Unfolding steps of I27 serve as a fingerprint signal to identify the correct single protein tether.
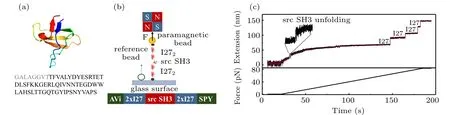
Fig.1.(a)The structure and amino acid sequence of protein src SH3(the grey letters show the eight N-terminus amino acids of unstructured polypeptide which is not showed in the structure). (b) Sketch of protein construct and single protein stretching experiment by magnetic tweezers. (c) Typical unfolding time trace obtained in force-ramp experiments with constant loading rate of 0.5 pN/s. Inset shows the unfolding step of src SH3.
4. Equilibrium folding and unfolding dynamics around critical force
As magnetic tweezers can maintain intrinsic constant force over long duration, equilibrium folding and unfolding dynamics studies can be easily carried out under constant forces close to the critical force of src SH3, which gives direct model-independent measurements of force-dependent dynamics. Figure 2(a)shows typical measurements of the folding and unfolding dynamics of src SH3 at constant forces of 4.5, 5.0, and 5.5 pN. The right panel shows the histogram of smoothed extension and Gaussian fitting with two peaks corresponding to unfolded and native state of src SH3,respectively.State with shorter extension is the native state,while that with longer extension is the unfolded state. This histogram clearly shows that protein has greater chance of staying at unfolded state with increasing stretching force.
Unfolding and folding probabilities as functions of time are obtained from cumulative distribution of lifetime of native state and unfolded state, respectively. The exponential fitting gives the corresponding unfolding ratekuand folding ratekfat each force(Figs.2(b)and 2(c)).
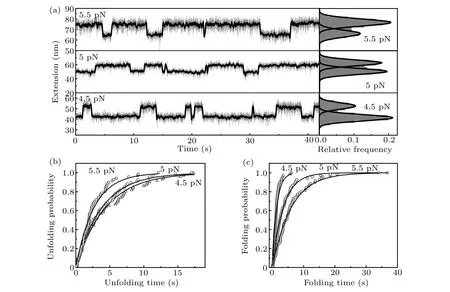
Fig.2. Equilibrium unfolding and refolding dynamics of src SH3 at constant forces. (a)Extension time courses of src SH3 were measured at constant forces of 4.5 pN,5 pN,5.5 pN.Corresponding relative frequencies of extension shown in the right panel were fitted with two-peak Gaussian functions.(b)and(c)Unfolding and folding probabilities of src SH3 at different forces as functions of time are obtained from cumulative distribution of lifetime of native state and unfolded state,respectively. Solid lines show exponential fitting curves to determine ku and kf of src SH3.
4.1. Force-jump measurement of unfolding rate
In order to explore the unfolding rate at higher force range,we performed the force-jump experiment from 4 pN to 11 pN(Fig. 3(a)). After one cycle of constant loading rate measurement, we applied small force of 0.5 pN for two seconds to let it fold to native state,then changed force to high values abruptly and maintained the same force for about 8 s(from 4 pN to 7 pN)and 5 s(from 8 pN to 11 pN)to record the unfolding step of src SH3(Fig.3(a)). Force-extension curve from constant loading rate measurement and the average extensions before and after the unfolding transitions in force-jump measurement are shown in Fig. 3(b). As is expected, the extensions of unfolded state in force-jump experiment are the same as the extension in constant loading rate experiment.
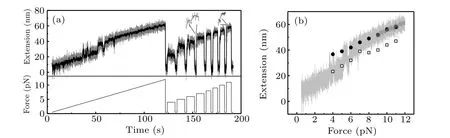
Fig.3. Force-jump measurement of the unfolding process. (a)Bottom panel shows the time course of force. Firstly,force increases from 0.5 pN to 12 pN with constant loading rate of 0.1 pN/s, then decreases to 0.5 pN abruptly and maintain for 2 s. After that, force jumps between high forces in the range of 4-11 pN and low force of 0.5 pN. Top panel shows the extension time course, which demonstrates the unfolding step. The same stretching processes are repeated 64 times. From the life time of native state at each force value, unfolding rates are obtained. (b)Force-extension curve obtained from force-ramp experiment of Fig. 3(a) (grey solid line) is shown together with extensions before (open squares) and after (open circles)the unfolding transition in force-jump experiment. Dark solid line shows the smoothed curve over five-second time windows.
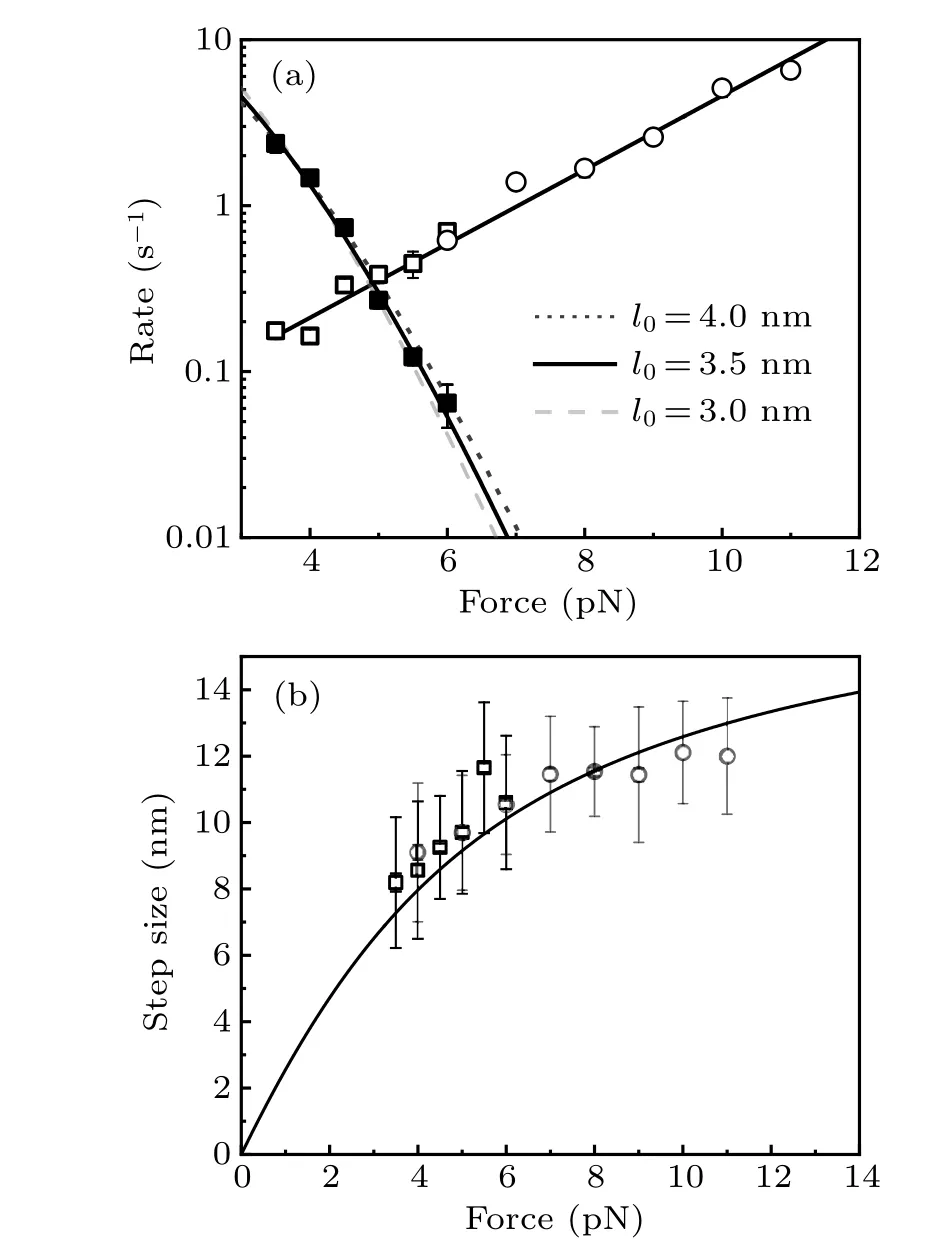
Fig. 4. Force-dependent folding and unfolding rates and unfolding step sizes of src SH3. (a)Folding rates(solid squares)and unfolding rates(open squires)of src SH3 were obtained from equilibrium constant force measurements, while unfolding rates (open circles) were obtained from force-jump experiment. The folding rates were fitted using Arrhenius’ law to estimate the size of folding transition state of 3.5±0.5 nm, while Bell’s model with xu=2.1±0.1 nm fits the force-dependent unfolding rate well. (b) Unfolding step sizes of src SH3 are obtained from equilibrium measurement(open squares) and force-jump measurement (open circles). Error bar is the standard deviation. Black curve is the theoretical curve of extension difference between unfolded polypeptide and native state.

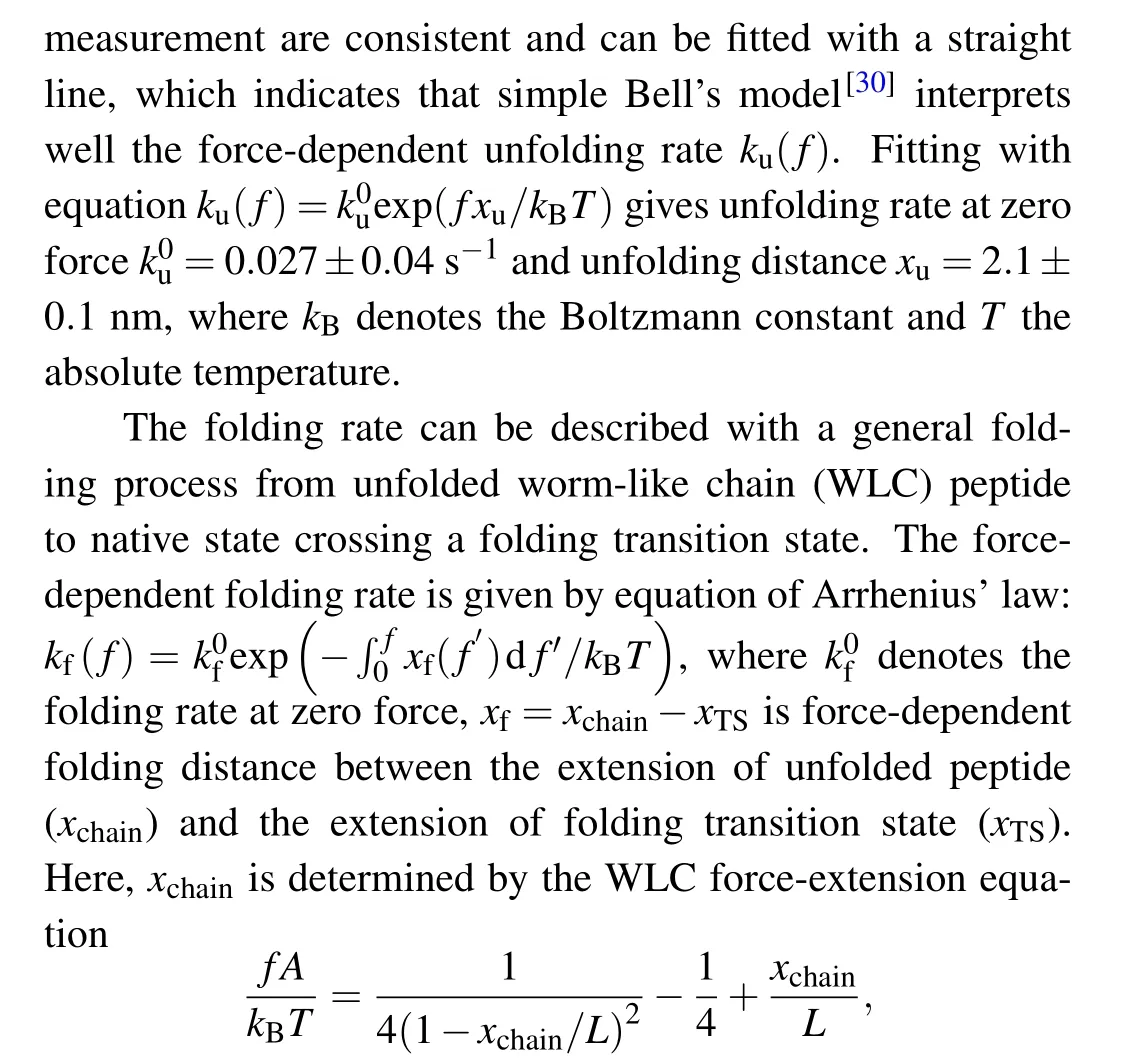
whereAis persistence length,Lthe contour length, andxTSthe extension of transition state. We suppose that the folding transition state is a specific conformation with orientational fluctuation, thenxTS=l0(coth(fl0/kBT)-kBT/fl0),wherel0is the N-C distance of this folding transition state.[17]Unfolded polypeptide has persistence lengthAof 0.8 nm and contour lengthLof 21.3 nm (0.38 nm per amino acid and 56 amino acids).[17,21]The fitting givesk0f=25 s-1andl0=3.5±0.5 nm.
Unfolding step sizes obtained from both equilibrium measurement and force-jump measurement are shown in Fig.4(b),which are consistent with the theoretical curve with contour length of unfolded peptideL=21.3 nm,persistence length of unfolded peptideA=0.8 nm,and the N-C distance of native state 0.64 nm.
5. Discussion and perspectives
Force-dependent unfolding rates show a perfect linear relationship with force when rates are plotted in logarithmic scale (Fig. 4(a)). The unfolding distancexuis about 2.1 nm over force range from 4 pN to 11 pN as obtained from the fitting of the force-dependent unfolding rate by Bell’s model.By adding the N-C distance of native state of 0.64 nm,the extension of unfolding transition state is about 2.74 nm. Forcedependent folding rates determine that the folding transition state has N-C distance of about 3.5 nm, from which the extension of folding transitionxTSis from 2.5 to 2.8 nm in force range of 4-6 pN, similar to the extension of unfolding transition state. Therefore, it indicates that the folding transition state is the same as the unfolding transition state, and there is a single pathway to between the native state and unfolded polypeptide.
Force-dependent folding free energy ΔG(f) =kBTln(kf(f)/ku(f)). As the lowest force in our measurement is smaller than 4 pN,the extrapolated value of zero force unfolding and folding ratesk0uandk0fmust be very close to the real value. Fromk0uandk0f, folding free energy at zero force ΔG(0) = 6.8kBT, which is consistent with biochemical measurement.[22]At zero force, if we suppose that the intrinsic transition ratek*=106s-1, then the unfolding free energy barrier at zero force can be calculated by the equationk0u=k*exp(-ΔG‡), which gives ΔG‡= 17.4kBT. Therefore, the folding free energy barrier is about 10.6kBT. For an unfolded polypeptide of 56 amino acids, the root-meansquare N-C distance of random coil can be estimated to be about 6 nm from free joined chain model with Kuhn length of 1.6 nm (twice of persistence length 0.8 nm). Therefore, the transition state locates at position in the middle of native state and unfolded polypeptide if we choose N-C distance as the reaction coordinate(Fig.5).[26,31,32]
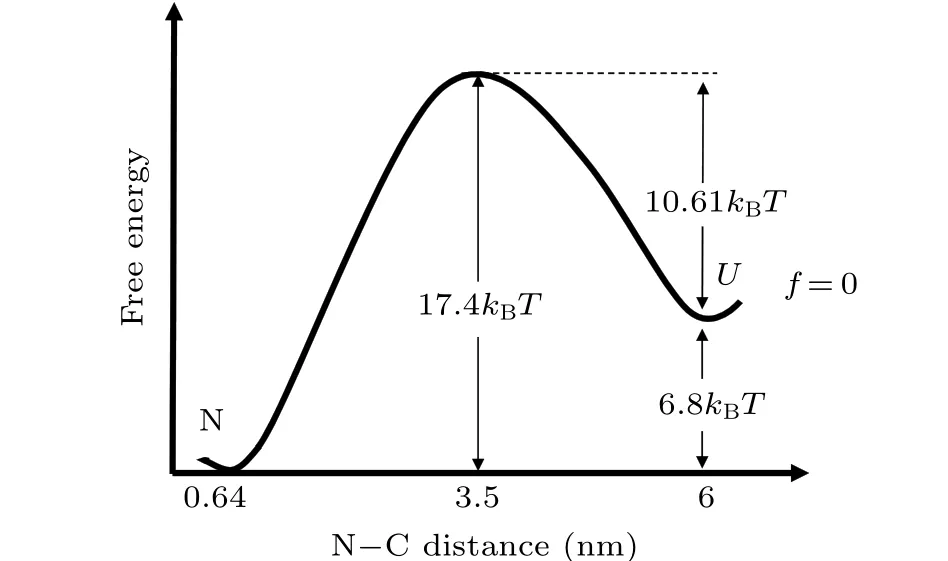
Fig. 5. Free energy landscape of src SH3 at zero force (solid line) is constructed with N-C distance as the reaction coordinate. Folding free energy,unfolding barrier,folding barrier,and location of the transition state are all quantified and marked.
Among single molecular manipulation techniques of AFM, optical tweezers, and magnetic tweezers, magnetic tweezers are most suitable to study the equilibrium folding and unfolding dynamics of proteins close to the critical force. Because critical forces of most proteins are smaller than 10 pN,the extrapolated results of zero force properties will have little deviation from the real value. Further temperature-dependent and denaturant-dependent measurement can be readily incorporated into magnetic tweezers experiments. We believe that more proteins with different topological structures and compositions of secondary structures will be studied by magnetic tweezers, and general protein folding mechanism will be revealed.
猜你喜欢
杂志排行
Chinese Physics B的其它文章
- Numerical simulations of partial elements excitation for hemispherical high-intensity focused ultrasound phased transducer*
- Magnetic-resonance image segmentation based on improved variable weight multi-resolution Markov random field in undecimated complex wavelet domain*
- Structure-based simulations complemented by conventional all-atom simulations to provide new insights into the folding dynamics of human telomeric G-quadruplex*
- Dual-wavelength ultraviolet photodetector based on vertical(Al,Ga)N nanowires and graphene*
- Phase-and spin-dependent manipulation of leakage of Majorana mode into double quantum dot*
- Deep-ultraviolet and visible dual-band photodetectors by integrating Chlorin e6 with Ga2O3
