Identification and tissue distribution of odorant binding protein genes in Harmonia axyridis (Coleoptera:Coccinellidae)
2021-03-26QUChengWANGRanCHEWunanLIFengqiZHAOHaipengWEIYiyunLUOChenXUEMing
QU Cheng,WANG Ran,CHE Wu-nan,LI Feng-qi,ZHAO Hai-pengWEI Yi-yun,LUO Chen,XUE Ming
1 College of Plant Protection,Shandong Agricultural University,Tai’an 271018,P.R.China
2 Institute of Plant and Environment Protection,Beijing Academy of Agriculture and Forestry Sciences,Beijing 100097,P.R.China
3 Department of Pesticide Sciences,Shenyang Agricultural University,Shenyang 110866,P.R.China
Abstract The olfactory system of insects is crucial in modulating behaviors such as host seeking,mating,and oviposition. Odorantbinding proteins (OBPs) are involved in semiochemical recognition. OBPs recognize and bind odorants and transport them to odorant receptors located in olfactory neurons. Harmonia axyridis (Coleoptera:Coccinellidae) is a widely used predacious biological control agent for many agricultural and forestry pests. This study identified 19 OBPs in H.axyridis based on the antennal and whole-body transcriptomes of adults and obtained all the full-length open reading frames,including 11 ‘Classic’ OBPs,7 ‘Minus-C’ OBPs and 1 ‘Plus-C’ OBP. They encoded 125 to 241 amino acid proteins with molecular weights ranging from 13.75 to 27.75 kDa and isoelectric points ranging from 4.15 to 8.80. Phylogenetic analyses were used to study the relationships between H.axyridis OBPs and OBPs from other species of Coleoptera. Quantitative real-time PCR (qPCR) analysis showed that HaxyOBP2,3,5,8,10,12,13,14,and 15 were highly expressed in antennae of both adult females and males. Moreover,HaxyOBP2,3,5,12,and 15 were more abundantly expressed in antennae than other body parts,while HaxyOBP13 and HaxyOBP14 were expressed predominantly,and at similar levels,in the head and antennae. The other OBP genes were highly expressed in non-olfactory tissues including the thorax,abdomen,legs,and wings. These results provide valuable information for further study of H.axyridis olfaction,which may ultimately enhance its use as a biocontrol agent.
Keywords:odorant-binding proteins,Harmonia axyridis,expression profile,phylogenetic tree
1.Introduction
The olfactory system of insects is important in regulating behaviors such as host location,mating,and oviposition (Pelosiet al.2006;Asahinaet al.2008). The main proteins of the insect olfactory system include odorant-binding proteins (OBPs),chemosensory proteins (CSPs),odorant receptors (ORs),ionotropic receptors (IRs),sensory neuron membrane proteins (SNMPs),and odorant degrading enzymes (ODEs) (Leal 2013;Pelosiet al.2014;Hickneret al.2016). In addition,recent studies showed that a new transport protein Niemann-Pick type C2 (NPC2),a type of small soluble protein involved in lipid metabolism and triglyceride accumulations,plays similarly crucial roles as OBPs and CSPs in chemical communication of insects,such as the worker Japanese carpenter antCamponotusjaponicus,the cotton bollwormHelicoverpaarmigerain,and the parasitoid waspMicroplitismediator(Ishidaet al.2014;Pelosiet al.2014;Zhenget al.2018;Zhuet al.2018). OBPs are involved in the initial steps of semiochemical recognition and play a key role in the olfactory system. They recognize and bind odorants that have entered the olfactory sensilla and transport them to odorant receptors located in the dendrite membrane of olfactory neurons (Zhouet al.2010;Daniet al.2011;Vieira and Rozas 2011).
Based on structural features and protein sequence similarity,insect OBPs can be divided into three major subclasses:‘Classic’ OBPs with six cysteine (Cys) residues,‘Minus-C’ OBPs with two less Cys residues,and ‘Plus-C’ OBPs with a Pro residue and two additional Cys residues (Zhouet al.2004;Schultzeet al.2012;Spinelliet al.2012). Since the OBP family was first reported inAntheraeapolyphemus(Vogt and Riddiford 1981),many OBPs have been gradually revealed by transcriptome analyses of Hemiptera,Lepidoptera,Hymenoptera,Neuroptera and Coleoptera. For example,15 OBPs were identified inIpstypographus(Anderssonet al.2013),31 inDendroctonusponderosae(Anderssonet al.2013),25 inHolotrichiaparallela(Juet al.2014),12 inChrysoperlasinica(Liet al.2015),28 inAthetislepigone(Zhanget al.2017b),8 inBemisiatabaci(Wanget al.2017b),and 41 inSitophiluszeamais(Tanget al.2019).
Harmoniaaxyridis(Coleoptera:Coccinellidae) is a predatory insect,widely distributed in Asia. It is a natural enemy of several agricultural and forestry pests and has promise as a biological control agent (Kochet al.2003;Wanget al.2015),therefore it was introduced to Europe,Africa and Oceania for pest management (Trouveet al.1997;Koch 2003). Predatory insects likeH.axyridis,similar to the well-studied phytophagous insects,use olfaction to locate hosts;however,their olfactory system is largely unexplored. OBPs,as the main protein of insect olfactory system,are a key element in prey searching behavior.
This study identified and analyzed OBP genes from the antennal and whole-body transcriptome datasets ofH.axyridis. The theoretical physicochemical parameters of the OBPs were characterized. The similarity and phylogeny of these OBPs and those available from other Coleoptera were analyzed. The tissue expression profiles of these OBP genes were determined by quantitative real-time PCR (qPCR). The results facilitate the identification of olfactory receptive mechanisms and provide valuable information for subsequent functional studies.
2.Materials and methods
2.1.Insects and tissues
Harmonia axyridiswere purchased from Beijing Kuoye Tianyuan Biological Technology Co.,Ltd.,China.Insects were reared in a growth chamber in Beijing Academy of Agriculture and Forestry Sciences at (23±1)°C with a 16 h L:8 h D photoperiod and 70% relative humidity,and fed withAphis craccivoraKoch (Hemiptera:Aphididae). Tissues including antennae (for transcriptome and tissue expression analyses),heads without antennae,thoraxes,abdomens,legs and wings of 3-5 day old adults of both sexes (for differential tissue expression analyses) were collected.Three biological replicates were conducted. The samples were stored at -80°C until use.
2.2.Identification of OBP genes and phylogenetic analysis
The transcriptome data sets of antennae and whole body were sequenced on the Illumina HiSep™ 4000 platform. Putative OBP genes were identified by searching the transcriptome database with keywords (odorant binding protein) and BLAST,and confirmed based on Blastx searches against the nonredundant GenBank database.Open reading frames (ORFs) were predicted in the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). All the ORFs obtained by PCR utilized the following conditions:1.0 μL template of cDNA,8 μL PremixTaq(TaKaRaTaq™ ver.2.0 plus dye,TaKaRa,Japan),1.0 μL forward and reverse primers (10 μmol mL-1) and 9 μL sterile water,for a total reaction volume of 20 mL PCR. The PCR reaction was started at 94°C for 3 min and followed by 35 cycles of 94°C for 40 s,52°C for 50 s and 72°C for 50 s,with a final hold at 72°C for 10 min in a Bio-Rad thermocycler (Bio-Rad DNA Engine Peltier Thermal Cycler,Bio-Rad,USA). The primers used are indicated in Appendix A.
Agarose gel electrophoresis was conducted with 1% agarose gels. The products were purified according to their expected sizes using the Wizard DNA Purification System (Promega,WI,USA) and cloned into a pMD18-T vector (TaKaRa,Japan). All the sequencing was performed by Sunbiotech (Beijing,China). The signal peptides were determined using the SignalP 5.0 Server (http://www.cbs.dtu.dk/services/SignalP/). ExPASy Proteomics Server (http://cn.expasy.org/tools/pi_tool.html) was used to compute the isoelectric point and molecular weight of the deduced protein sequences. The cDNA sequence assembly and multiple sequence alignment were performed with DNAMAN 5.2.2 (Lynnon BioSoft).
Phylogenetic analysis of OBPs was performed based on amino acid sequences contained in reports of OBPs of Coleoptera. The phylogenetic tree was constructed using the MEGA5.05 Program with the nearest neighbor-joining method supported by 1 000-fold bootstrap resampling.
2.3.Tissue expression pattern of OBP genes by qPCR
Total RNA was extracted from these tissues using the TRIzol reagent (Invitrogen,Carlsbad,CA,USA) following the manufacturer’s instructions. The quality and quantity of RNA samples were assessed with a Thermo Scientific NanoDrop 2 000 UV-Vis spectrophotometer (Thermo Fisher Scientific Inc.,Waltham,MA,USA). The cDNA was synthesized by the PrimeScriptTMRT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa,Japan) according to the manufacturer’s protocol. qPCR was carried out in 20 μL reactions containing 2.0 μL cDNA,10 μL SYBR PremixExTaqTM II (TaKaRa,Japan),1 μL forward primer (10 μmol L-1),1 μL reverse primer (10 μmol L-1),0.4 μL Rox Reference Dye II (TaKaRa,Japan) and 5.6 μL nuclease free water using an ABI PRISM 7500 qRT-PCR System (Applied Biosystems,USA). The amplification conditions for qPCR were as follows:95°C for 30 s,followed by 40 cycles of 95°C for 5 s,and 60°C for 34 s. All test samples were performed in triplicate. Primers for qPCR were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primerblast/index.cgi?LINK_LOC=BlastHome) and listed in Appendix B. The EF1A and RPS13 were used as endogenous control genes to normalize the expression levels of OBP genes (Quet al.2018). Relative quantification was performed using the 2-ΔΔCTmethod (Schmittgen and Livak 2008). One-way ANOVA (SPSS 19.0,Chicago,IL,USA) followed by Tukey test was used to compare the relative fold change of genes in different issues.
3.Results
3.1.Identification of OBPs in H.axyridis
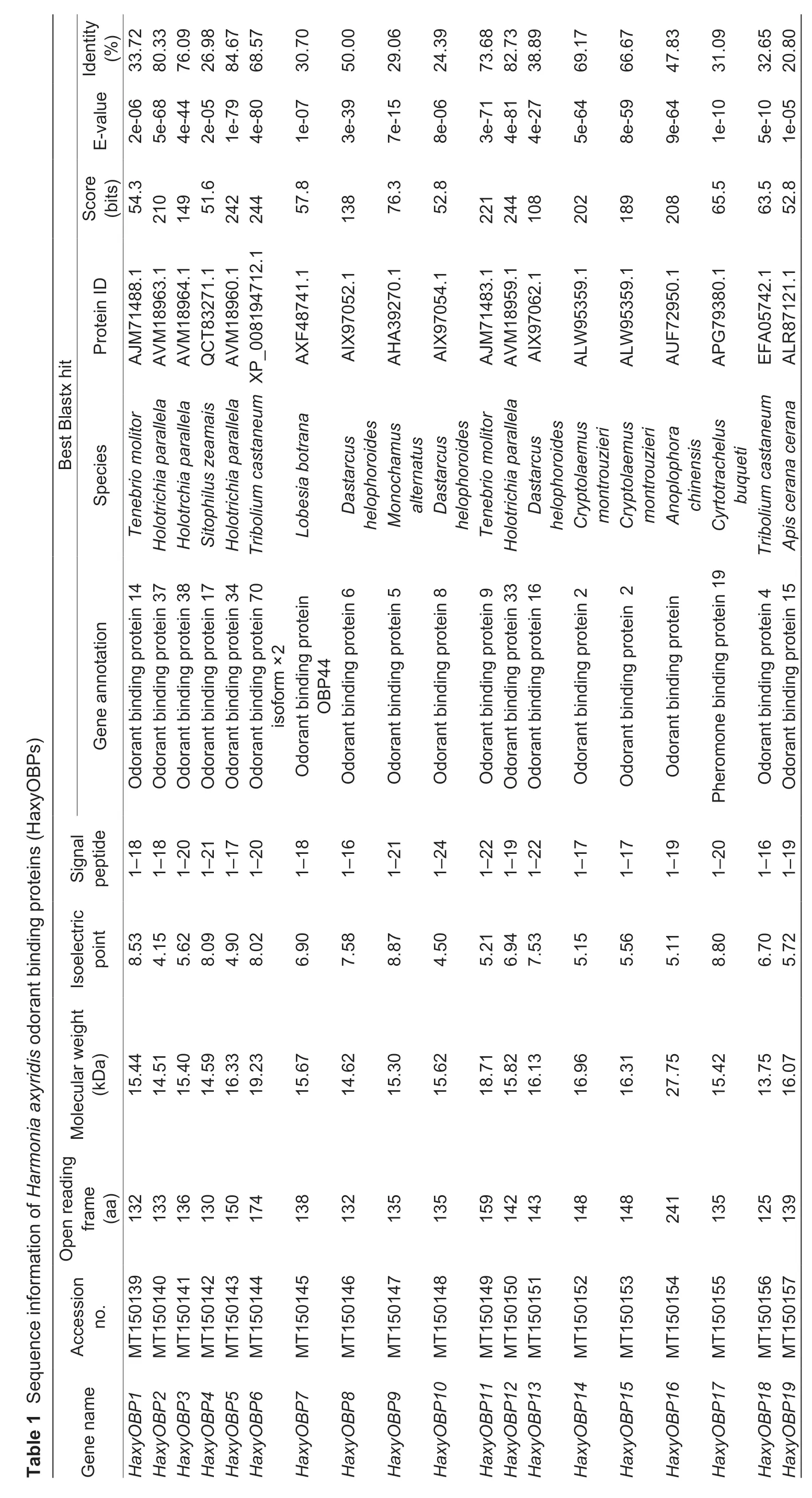
A total of 19 OBP genes were obtained by searching the antennal and whole-body transcriptome ofH.axyridis,henceforth namedHaxyOBP1-19(Table 1). Genes were successfully cloned and contained full-length ORFs encoding 125 to 241 amino acids. Each of the OBPs had a signal peptide that varied from 16 to 24 amino acid residues,with molecular weights ranging from 13.75 to 27.75 kDa and isoelectric points of 4.15-8.80 (Table 1). Based on the number and location of the conserved cysteines,the 19 HaxyOBPs were divided into three subfamilies:Classic OBPs includingHaxyOBP2,4,5,6,10,11,12,13,14,15and18,which had six conserved cysteines,‘Plus-C’ OBPsHaxyOBP16which contained two additional conserved cysteines and a conserved proline immediately after the sixth cysteine (Fig.1),and ‘Minus-C’ OBPs includingHaxyOBP1,3,7,8,9,17and19which had four conserved cysteines with C2 and C5 missing (Fig.2).

Fig.1 Alignment of Harmonia axyridis ‘Classic’ odorant-binding proteins (OBPs;HaxyOBP2,4,5,6,10,11,12,13,14,15 and 18) and ‘Plus-C’ OBP (HaxyOBP16). The conserved cysteine residues are indicated with * under the sequence.

Fig.2 Alignment of Harmonia axyridis ‘Minus-C’ odorant-binding proteins (OBPs). The conserved residues in the ‘Minus-C’ OBPs motif are indicated with * under the sequence.
3.2.Phylogenetic analysis of H.axyridis OBPs
A phylogenetic analysis of OBPs was conducted using 145 OBP protein sequences from eleven species of Coleoptera.The phylogenetic tree showed that the 19 HaxyOBPs shared high similarity with the sequences of other known Coleoptera OBPs and were clustered together within the same branches. These OBPs were grouped into three classes:the Classic OBPs,the ‘Minus-C’ OBPs and the ‘Plus-C’ OBPs (Fig.3). This suggested that the variation in molecular structure might be the result of the proteins having different roles in other biological processes.
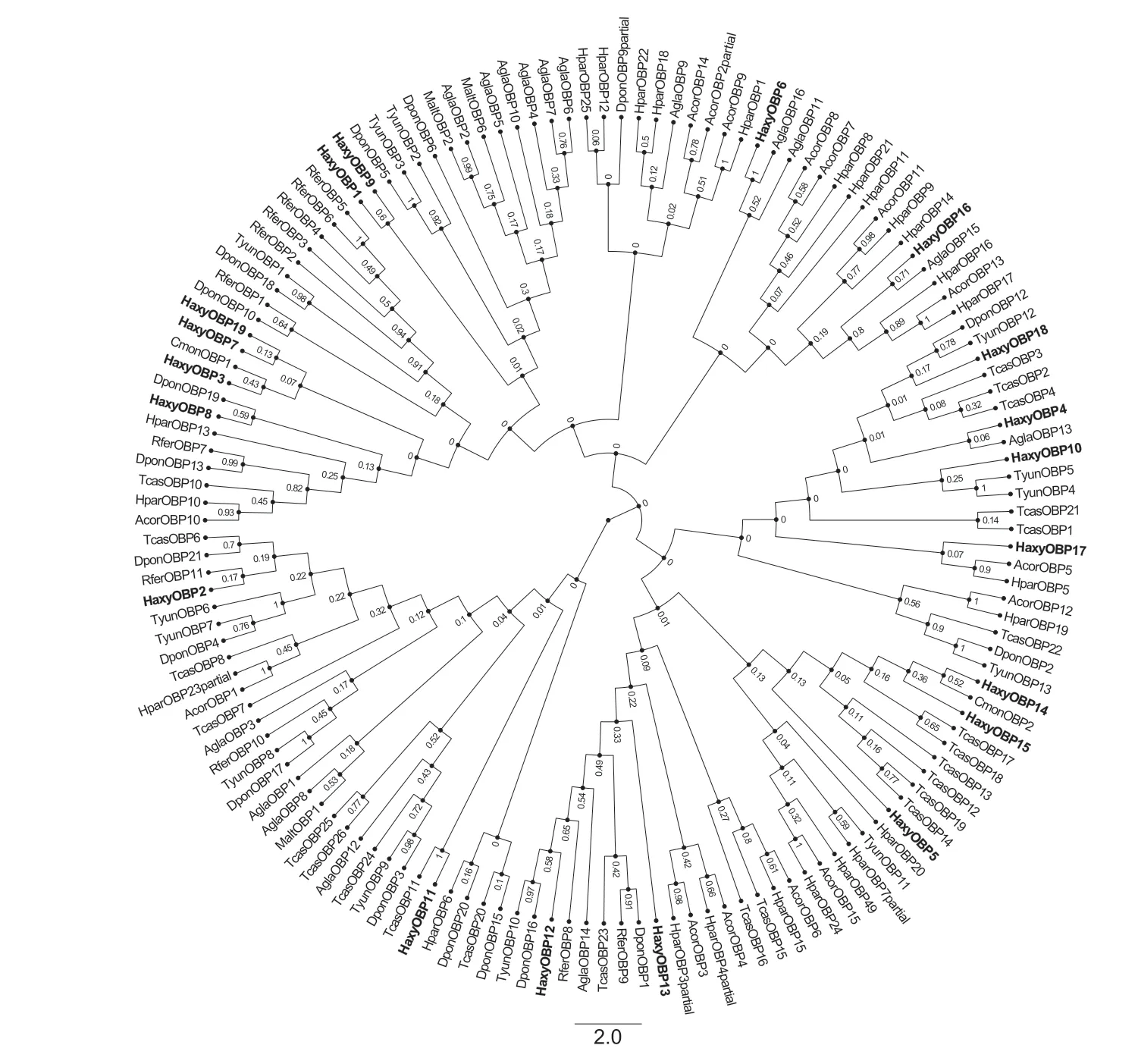
Fig.3 Neighbor-joining tree of candidate odorant-binding proteins (OBPs) identified from Harmonia axyridis compared with other Coleopteran OBPs. Sequences include Monochamus alternatus (Malt),Anomala corpulenta (Acor),Cryptolaemus montrouzieri (Cmon),Anoplophora glabripennis (Agla),Dendroctonus ponderosae (Dpon),Rhynchophorus ferrugineus (Rfer),Tomicus yunnanensis (Tyun),Holotrichia parallela (Hpar) and Tribolium castaneum (Tcas). The tree was constructed using MEGA5.05,and values at nodes are bootstrap values based on 1 000 re-sampling points. OBPs identified from H.axyridis are shown in bold.
3.3.Tissue expression pattern of the H.axyridis OBPs
qPCR was used to determine the relative expression levels of all putative OBPs in different tissues,including the antennae,heads without antennae,thoraxes,abdomens,legs,and wings of adult females and males. The results indicated that all 19 putative OBP genes had significantly different expression levels in different tissues (P<0.05).HaxyOBP2,3,5,8,10,12,13,14,and15were highly expressed in antennae of both adult females and males. In addition,five OBP genes (HaxyOBP2,3,5,12,and15) were significantly more highly expressed in the antennae compared to other body parts (P<0.05).HaxyOBP13and14were most dominantly expressed in the head and antennae,whileHaxyOBP4was highly expressed in the head.HaxyOBP17was highly expressed in the abdomen.HaxyOBP1,6,7,9,11,18and19showed high expression in nearly all tissues (Fig.4).
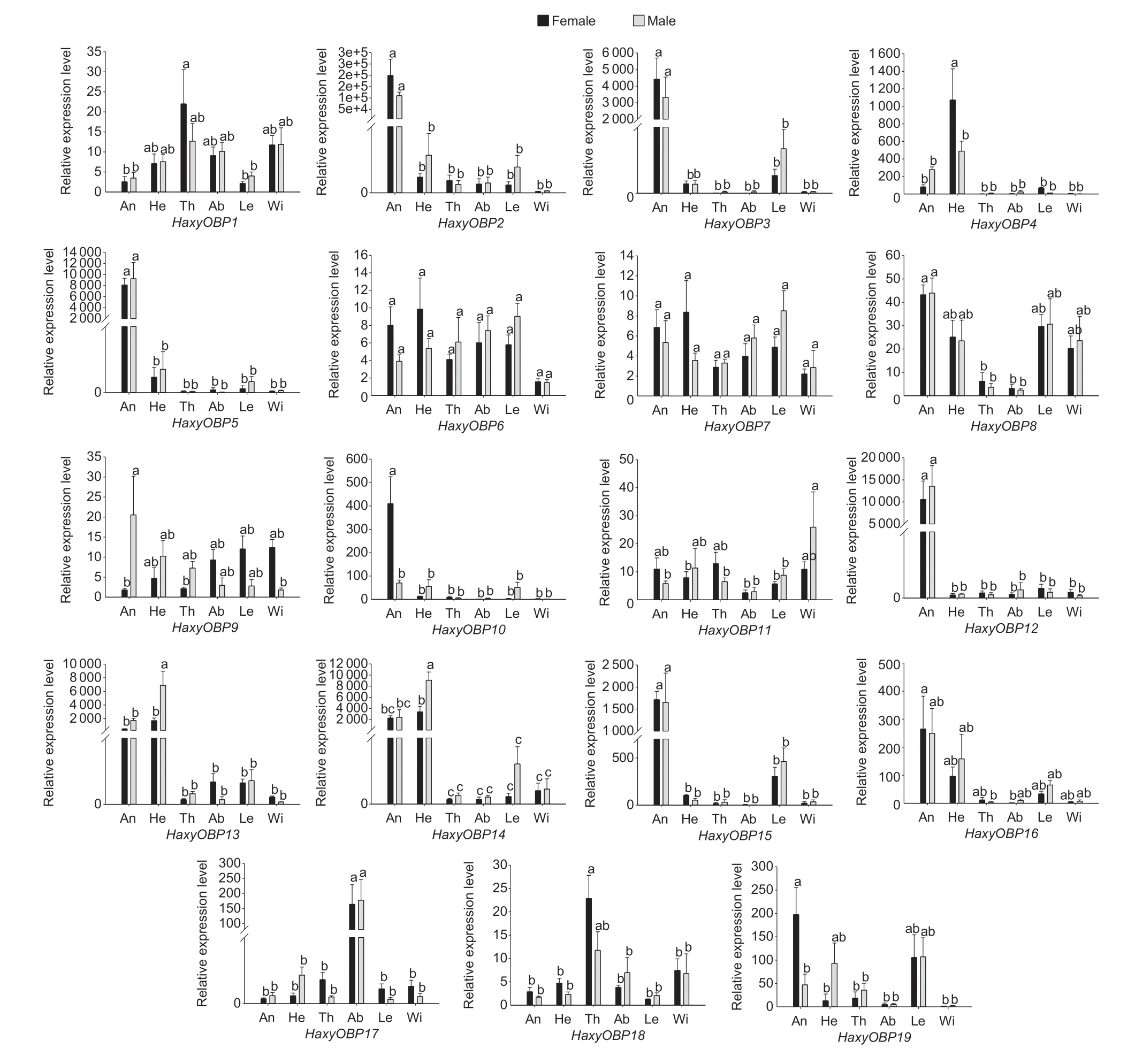
Fig.4 Relative expression levels of HaxyOBP1-19 genes in different tissues of both female and male adults. An,antennae;He,head;Th,thorax;Ab,abdomen;Le,leg;Wi,wing. The EF1A and RPS13 were used as reference genes to normalize expression levels in each cDNA sample. Relative expression levels are indicated as mean±SEM of three biological replicates. The comparisons were analyzed using one-way ANOVA,and the different letters above each bar indicate significant differences (Tukey test,P<0.05).
4.Discussion
OBPs are involved in transferring odor molecules to odor receptors (ORs),which play a key role in an insect’s successful perception of sex pheromones,alarm pheromones,and host plant volatiles (Pelosiet al.2006;Britoet al.2016). The present study identified 19 OBPs ofH.axyridisby searching the antennal and whole-body transcriptome,and named them byHaxyOBP1-19. OBP genes have also been identified from other Coleoptera,including 15 OBPs ofI.typographus(Anderssonet al.2013),49 OBPs ofTriboliumcastaneum(Dippelet al.2014),21 OBPs ofDendroctonusvalens(Guet al.2015),26 OBPs ofColaphellusbowringi(Liet al.2015),10 OBPs ofLissorhoptrusoryzophilus(Yuanet al.2016),21 OBPs ofCallosobruchuschinensis(Zhanget al.2017a),28 OBPs ofEucryptorrhynchusbrandti(Wenet al.2018),and 31 OBPs ofE.scrobiculatus(Wenet al.2018). The number of HaxyOBPs identified in this study is similar with these previous reports. However,OBPs alone are insufficient for comprehensive understanding of insect olfaction,and other olfactory proteins require further study.
It is widely regarded that OBPs located in different tissues may have different functions in insects (Pelletier and Leal 2009;Xuet al.2010;Mitakaet al.2011;Wanget al.2017a). The tissue expression pattens in this study may provide evidence for the potential functions of these genes. Antennae are the most important olfactory organs in insects,and the highly expressed genes in olfactory organs are likely to play a role in the antennal chemical recognition processes (Cuiet al.2019). The results showed thatHaxyOBP2,3,5,12,and15were significantly more highly expressed in the antennae compared to other body parts,which may be related to the antennal recognition processes that trigger forging,mating and other behaviors inH.axyridis. Besides,HaxyOBP13and14were highly expressed in both the antennae and head without antennae. Heads contain chemosensory organs such as maxillary palps and the proboscis in addition to antennae. OBP expression in the proboscis/maxillary palp sensilla is associated with gustatory responses (Shanbhaget al.2001;Del Campoet al.2011). Some HaxyOBPs were expressed predominantly in the thorax,wings,and abdomen,suggesting that these OBPs may participate in other physiological functions. For example,HarmOBP10,which is highly expressed in the male seminal fluid ofH.armigeraand carries oviposition deterrents,can be transferred to females and mark fertilized eggs. The OBP prompts female adults to fly away from the location where the first egg was laid (Sunet al.2012).
It has been reported that OBPs may have different expression levels between males and females (Yanet al.2016;Caoet al.2018;Songet al.2018). This study found that HaxyOBP in males and females were dissimilar.HaxyOBP4and9were significantly more highly expressed in male antennae than in female antennae inH.axyridis. This is evidence thatHaxyOBP4and9may be significant in male chemoreception that is related to mediating behaviors like mating,foraging and avoidance,and may play a role similar to the pheromone binding protein (PBP) (Gonget al.2014). TheHaxyOBP6,10and19genes were expressed at much higher levels in female antennae than in male antennae,suggesting thatHaxyOBP6,10and19may be involved in identification of prey volatiles or suitable sites for oviposition (Matsuoet al.2007). The expression patterns will be helpful for understanding OBPs inH.axyridisand might facilitate their future functional characterization.
5.Conclusion
The work presented here brings an identification of 19 OBPs from the antennal and whole-body transcriptome ofH.axyridis,and the tissue expression profiles of these genes were also constructed. Tissue distribution of the OBP genes showed that nearly half of them (HaxyOBP2,3,5,8,10,12,13,14,and15) were highly expressed in the antennae of both sexes. Other OBP genes were also enriched in nonolfactory tissues including the thorax,abdomen,legs,and wings. These results provide the basis for further research on the functions of the OBPs ofH.axyridis,which might lead to novel pest control strategies.
Acknowledgements
This work was supported by the National Key Research and Develop Program of China (2017YFD0200400),the Shandong Province Modern Agricultural Technology System Peanut Innovation Team,China (SDAIT-04-08) and the Beijing Leafy Vegetables Innovation Team of Modern Agroindustry Technology Research System,China (BAIC07-2020).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
