Karlodinium elegans sp. nov. (Gymnodiniales, Dinophyceae), a novel species isolated from the East China Sea in a dinoflagellate bloom*
2021-02-22JingyiCENJianyanWANGLifenHUANGYarouLINGuangmaoDINGYuzaoQISonghui
Jingyi CEN , Jianyan WANG , Lifen HUANG , Yarou LIN , Guangmao DING , Yuzao QI , Songhui LÜ ,
1 Research Center of Harmful Algae and Marine Biology, Jinan University, Guangzhou 510632, China
2 Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai 519000, China
3 Department of Science Research, Beijing Museum of Natural History, Beijing 100050, China
4 Fujian Fishery Resources Monitoring Center, Fuzhou 350003, China
5 Key Laboratory of Eutrophication and Red Tide Prevention of Guangdong Higher Education Institutes, Jinan University, Guangzhou 510632, China
Abstract On May 24-29, 2019, a harmful algal bloom occurred in Pingtan coastal areas, Fujian, southeast China, and caused mass mortality of cage-cultured fish. Two clonal cultures of an unknown naked dinoflagellate were set up from seawater samples taken during the bloom. The cultures were examined for morphological features, ultrastructure characters, photosynthetic pigments, the large subunit (LSU) of the rRNA gene, and the internal transcribed spacer (ITS) sequences, as well as acute toxicity analysis. The cell was unarmored, small-sized, and ovoid, and was characterized by elaborate striations on the epicone and hypocone. The nucleus was large, ellipsoid to oval or kidney-shaped, and centrally located in the cell. A long linear apical groove originated above the sulcus in the ventral epicone and extended to the dorsal side. An elongate, slit-like “ventral pore” was located on the left of the epicone, well away from the apical groove. The chloroplasts were yellowish brown, numerous, band like, and irregularly distributed in the cell periphery. Fucoxanthin was the main accessory pigment composition. Phylogeny topology reconstructed on partial LSU rDNA showed that the unknown dinoflagellate branched as a sister species to Karlodinium sp. (strain IFR981 & IFR797, from France) and Karlodinium corrugatum (strain KDGSO08, from Australia), with genetic divergences of 0.6% and 3.3%, respectively. Based on the morphological and molecular phylogenetic analysis, we describe the novel dinoflagellate as Karlodinium elegans sp. nov. A toxicity assay revealed that the clonal culture of K. elegans (strain PTB601) had no adverse effect on brine shrimp ( Artemia salina) and marine medaka ( Oryzias melastigma), indicating it may not be a toxic species.
Keyword: Karlodinium; Kareniaceae; harmful algal bloom; toxicity; dinoflagellate
1 INTRODUCTION
The genus Karlodinium belongs to the family Kareniaceae, and was established by Daugbjerg in 2000 based on a comprehensive analysis of morphological characteristics, phylogenetic data, and photosynthetic pigment composition (Daugbjerg et al., 2000; Bergholtz et al., 2006). Key morphological features of Karlodinium species are the presence of a straight apical groove and a ventral pore on the cell epicone (Daugbjerg et al., 2000). Another important characteristic is the unique type of amphiesma with plugs, which were observed under transmission electron microscopy (Bergholtz et al., 2006; Siano et al., 2009). However, recent studies revealed that the ventral pore is absent in some Karlodinium species, e.g. K. digitatum, K. zhouanum, K. jejuense, K. ballantinum, and K. antarcticum (Bergholtz et al., 2006; de Salas et al., 2008; Nézan et al., 2014; Li and Shin, 2018; Luo et al., 2018). In 2018, Luo et al. (2018) emended the definition of Karlodinium J. Larsen as unarmored dinoflagellates with chloroplasts, containing internal, lenticular pyrenoids, fucoxanthin or fucoxanthin derivatives as main accessory pigments, and a straight apical groove.
Fourteen species have been identified in the genus Karlodinium (Guiry and Guiry, 2020). However, biodiversity and biogeography of this group are supposed to be underestimated as Karlodinium species is small-sized, unarmored, and easily deformed after fixation for identification (Siano et al., 2009). Most species of Karlodinium are notorious for causing harmful algal blooms (HABs). Some species produce karlotoxins or other potentially toxic chemical compounds, and blooms may cause fish kills and endanger other marine life. Karlodinium veneficum, a species that produces karlotoxins with hemolytic, cytotoxic, and ichthyotoxic properties (Bachvaroff et al., 2008; Van Wagoner et al., 2008), has caused many HABs and fish kills around the world (Deeds et al., 2002; Wang et al., 2011; Adolf et al., 2015). Karlodinium australe was reported for having caused massive mortality of cage-farmed fishes in the West Johor Strait of Malaysia in February 2014 (Lim et al., 2014). Another toxic species, K. armiger , has caused mortality of bivalves, rotifers, copepods, and finfishes (Garcés et al., 2006; Rasmussen et al., 2017).
During May 24-29, 2019, a bloom dominated by K. digitatum and Prorocentrum donghaiense occurred in Pingtan coastal areas in Fujian, southeast (SE) China (Cen et al., 2020). An unknown naked dinoflagellate was isolated from water samples collected during monitoring of the bloom. In this study, we analyzed the cell morphology, molecular sequences, pigment composition, and toxicity of the dinoflagellate, on which we describe it as a novel species, Karlodinium elegans sp. nov.
2 MATERIAL AND METHOD
2.1 Specimen collection and cultivation

Fig.1 Map of sampling site
Two clonal cultures (PTB601 and PTB602) were created by isolating single cells from seawater samples collected during a bloom in Pingtan coastal area, Fujian, SE China (25°38′3.7″N, 119°41′42.7″E) (Fig.1) on May 29, 2019, and were maintained in L1 medium at 20 °C, salinity 27, and irradiance 80 μmol photons/(m2·s) provided by cool-white fluorescent lamps with a photoperiod of 12 h:12 h light:dark. The two strains were deposited at the Algal Culture Collection of the Research Center for Harmful Algae and Marine Biology, Jinan University, Guangzhou, China.
2.2 Light microscopy (LM)
Live cells were examined and photographed by Olympus BX61 microscope (Olympus, Tokyo, Japan) with a QImaging Retiga 4000R digital camera (QImaging, Surrey, BC, Canada). The chloroplast autofluorescence of live cells was observed on the Olympus BX61 microscope using a blue excitation device. For epifluorescence of nucleus, live cells were stained with SYBR Safe DNA (Thermo Fisher, Waltham, MA, USA) and photographed under an Olympus filter set (excitation: 360-370 nm; emission: 420-460 nm).
2.3 Cell measurement
Cell length, width and cingular displacement were measured on at least 40 randomly selected cells on a PC using Image-Pro Plus 6.0 image acquisition and analysis software (QImaging, Surrey, BC, Canada) on the Olympus BX61 light microscope.
2.4 Scanning electron microscopy (SEM)
Cells in the exponential growth phase were harvested and fixed with osmium tetroxide (OsO4) (a final concentration of 2%) and double-fixed with a final concentration of 1% acidic Lugol’s solution and 1% glutaraldehyde. Fixed cells were adhered to a cover slip coated with poly-L-lysine for 30 min. Samples were then dehydrated in a series of ethanol with concentrations of 10%, 30%, 50%, 70%, 90%, 95%, and three times in 100%, 10 min in each step. The samples were critical-point dried using a Leica EM CPD300 (Leica Microsystems, Mannheim, Germany) with liquid CO2. They were coated with gold-palladium and observed in a Zeiss Ultra 55 field emission scanning electron microscope (Zeiss, Jena, Germany).
2.5 Transmission electron microscopy (TEM)
Cells were double fixed with OsO 4 (a final concentration of 0.5%) and glutaraldehyde (a final concentration of 2%) for 30 min at room temperature, then centrifuged and rinsed 3 times using L1 culture medium. The collected pellet was post-fixed in 1% O s O4for 1 h at 4 °C, then washed quickly in distilled water and dehydrated in a series of ethanol (10%, 30%, 50%, 70%, and 90%, followed by three changes of 100%) for 10 min each, with a final dehydration in propylene oxide twice. The dehydrated materials were embedded in Epon resin and sectioned on an ultra-cut (UCT) ultra-microtome (Leica Microsystems, Wetzlar, Germany) using diamond knife. After being mounted on slot grids, the sections were stained with uranyl acetate and lead citrate, 10 min in each staining solution, and observed in a Jeol-1010 transmission electron microscope (Jeol Ltd., Tokyo, Japan).
2.6 Pigment analysis
An algal culture (strain PTB601) in mid-exponential phase was filtered onto a (nominal pore size of 1.2 μm) GF/C filter (Whatman, Maidstone, UK). Photosynthetic pigments were extracted according to Zapata et al. (2000). high-performance liquid chromatography (HPLC) analysis of pigment components followed the method described by Wang et al. (2018). Identification and quantitation of pigments were performed by comparison of UV-VIS absorption spectral properties and retention time to the authentic standards (DHI Inc., Copenhagen, Denmark).
2.7 DNA extraction and PCR amplification of LSU and ITS
About 2-mL cultures in the mid-exponential phase were centrifuged (2 000 r/min, 1 200× g) for 10 min at room temperature, and the pellets were stored at -80 °C until extraction. Genomic DNA was extracted using a MiniBEST Universal Genomic DNA Extraction Kit (Takara, Dalian, China) according to the manufacturer’s instructions, and was used as the DNA template for PCR reaction.
Partial LSU rDNA (D1-D3 region) was amplified using primers D1R (5′-3′: ACCCGCTGAATTTAAGCATA) (Scholin et al., 1994) and D3B (5′-3′: TCGGAGGGAACCAGCTACTA) (Nunn et al., 1996). ITS sequence was amplified with ITS1F (5′-3′: TCGTAACAAGGTTTCCGTAGGTG) and ITS1R (5′-3′: ATATGCTTAAGTTCAGCGGG) (Pin et al., 2001). PCR reaction was carried out in a PTC-200 Peltier Thermal Cycler (MJ Research, San Francisco, CA, USA) using amplification programs: 3 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 57 °C, 1 min at 72 °C, and a final extension of 6 min at 72 °C. The PCR products were sequenced by Beijing Genomics Institute (Guangzhou, China). New sequences were deposited in GenBank under accession numbers MT161377 and MT161378 (LSU rDNA), and MT161377 and MT161378 (ITS).
2.8 Sequence alignment and phylogenetic analysis
Adjusted sequences were aligned with other related Kareniaceae species downloaded from GenBank using ClustalX (Thompson et al., 1997). Gymnodinium catenatum was chosen as outgroup for LSU phylogeny and Heterosigma akashiwo as outgroup for ITS phylogeny. Maximum likelihood (ML) analyses were carried out using MEGA 7.0, and the model T3+G+I was selected. Node support was assessed with 1 000 bootstrap replicates (Tamura et al., 2013). Bayesian inference (BI) was carried out with MrBayes 3.1.2 following the best-fitting model (Ronquist and Huelsenbeck, 2003).
2.9 Acute toxicity analysis
Brine shrimp ( Artemia salina) bioassay: the toxic effect of the dinoflagellate was tested on the brine shrimp. The A. salina was incubated at 25 °C in a 500-mL glass beaker at 24 h. The dinoflagellate culture in the exponential growth phase was diluted to a concentration of 1.121×107cells/L; the L1 medium and P. donghaiense (3×107cells/mL) culture were used as the blank control and negative control, respectively. Ten specimens of A. salina were used in each test, and the test was performed in triplicate. Survival/mortality of A. salina was observed for 48 h and recorded every 3 h.
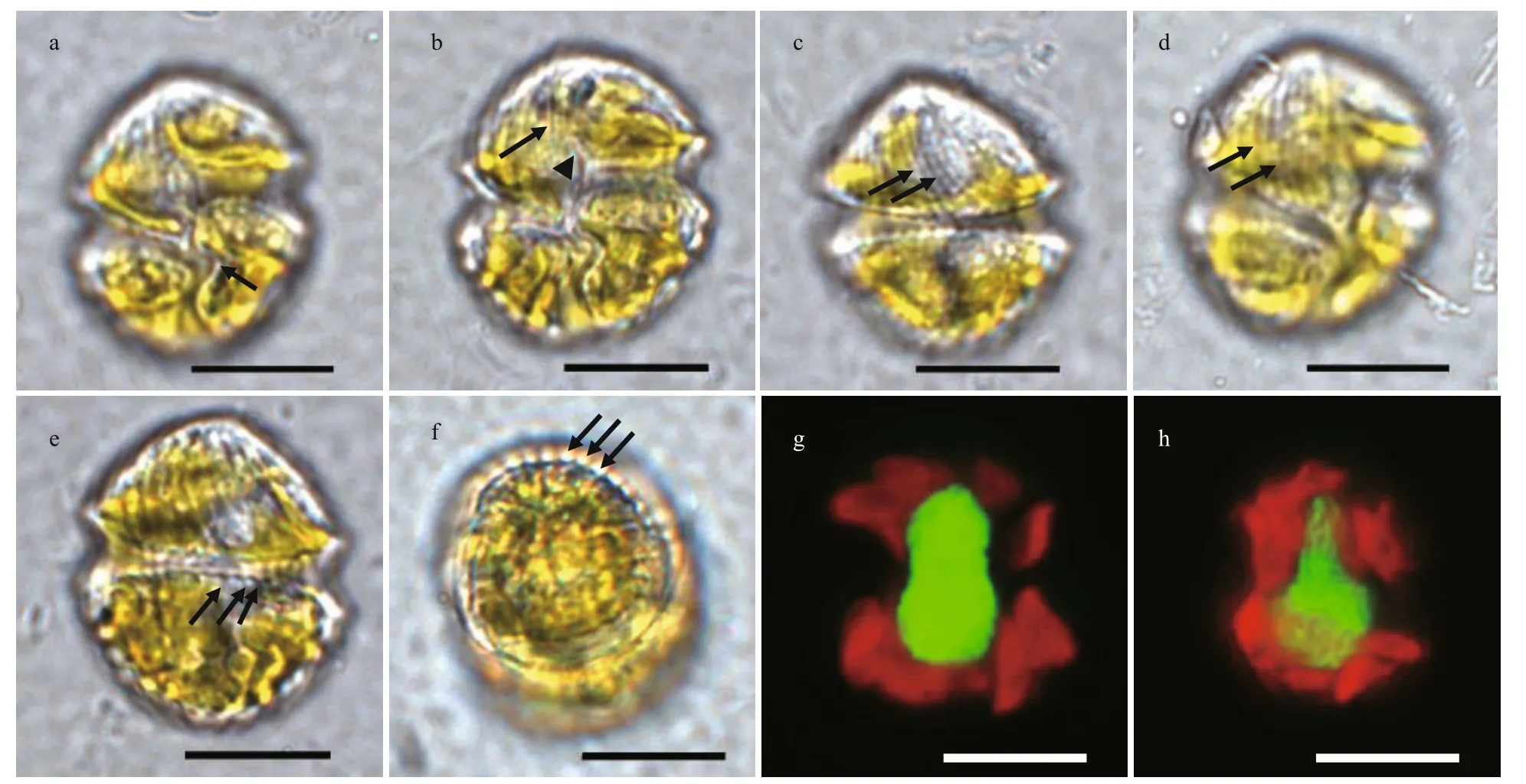
Fig.2 Light microscopy of vegetal cells of Karlodinium elegans strain PTB601
Marine medaka ( Oryzias melastigma) bioassay: Adult O. melastigma were kindly provided by the State Key Laboratory in Marine Pollution, City University of Hong Kong. Fish were cultured in seawater at 25 °C, in a photoperiod of 12 h:12 h light:dark, and fed twice daily. Fifteen healthy, 2-week old (after hatching) O. melastigma were transferred into a 500-mL glass beaker containing 300-mL algal culture. Acute toxicity of the dinoflagellates on O. melastigma was assessed by measuring the lethal effect in 48 h. Meanwhile, 300 mL of seawater and P. donghaiense culture (1×107cell/L) were used for the blank control and negative control, respectively. The survival of fish was observed after 0, 4, 8, 12, 24, and 48 h. Each set was performed in triplicate.
3 RESULT
Karlodinium elegans J. Y. Cen, S. H. Lu, & J. Y. Wang sp. nov. (Figs.2-8)
Diagnosis: Unarmored, ovoid in shape, 19-27 μm long and 15-23 μm wide. Epicone conical with parallel striations curving to the left side, a row of amphiesmal knobs ornamenting the right side of each stria. Hypocone hemispherical, decorated with longitudinal and horizontal stripes. Cingulum displaced 30% of the cell length. Sulcus extended onto the epicone as a finger-like intrusion. A tube-shaped structure in the intercingular region. Apical groove deep and straight, originating from the right of the sulcal intrusion and extending halfway down the dorsal side of the epicone. A slit-like “ventral pore” present. Nucleus large, kidney-shaped, centrally located in the cell from the epicone to the hypocone. Several yellowish-brown, band-like chloroplasts, each with an internal pyrenoid.
Holotype: The scanning electron microscopy stub containing cells of strain PTB601 was deposited as holotype at Beijing Museum of Natural History, Beijing 100050, China.
Type locality: Pingtan coastal areas of Fujian (25°38′3.7″N, 119°41′42.7″E), China. Collection date: 29 May, 2019.
GenBank accession number: MT161377 and MT161379, the nuclear-encoded LSU and ITS sequence of strain PTB601, respectively.
Etymology: Latin elegans, elegant, referring to the prominently elaborate striations on the cell epicone and hypocone.
Distribution: Pingtan coastal water, East China Sea.
3.1 Morphology of Karlodinium elegans Light microscopy (LM)
Strain PTB601 of K. elegans was unarmored, smallsized, with average cell length of 18.69-26.69 μm (22.87±1.50 μm, n=40) and average width of 14.69-23.05 μm (18.95±1.63 μm, n=40). The ratio of cell length to width was 1.12-1.37 μm (1.21±0.05 μm, n=40). General morphology features of K. elegans and related Karlodinium species are shown in Table 1.
Cells were ovoid in outline; the epicone was conical, and the hypocone was hemispherical and was truncated by the sulcus (Figs.2a & 5a). The sulcus was sigmoid, intruded onto the epicone as a finger-like structure, and was distinctively convex at the left lobe of hypocone (Figs.2a-b & 5a). The apical groove was elongated and straight, starting above the anterior end of the sulcus (Figs.2b & 5a). The cingulum was deeply incised, left handed, and equatorially located on the cell, with a displacement of ca. 30% of the cell length on the ventral side (Figs.2a, c, & 5a). Longitudinal and parallel striations were evenly distributed on the epicone, and curved to the left side, as seen in the dorsal view (Figs.2d & 5a). The surface of the hypocone was decorated with longitudinal stripes, arranged differently from those on the epicone (Figs.2e, f, & 5a-b). The nucleus was large, kidneyshaped, and centrally situated in the cell extending in both the epicone and the hypocone (Fig.2g & h). The yellowish-brown and band-like chloroplasts were distributed in the cell periphery, and surrounded the nucleus (Fig.2g & h).
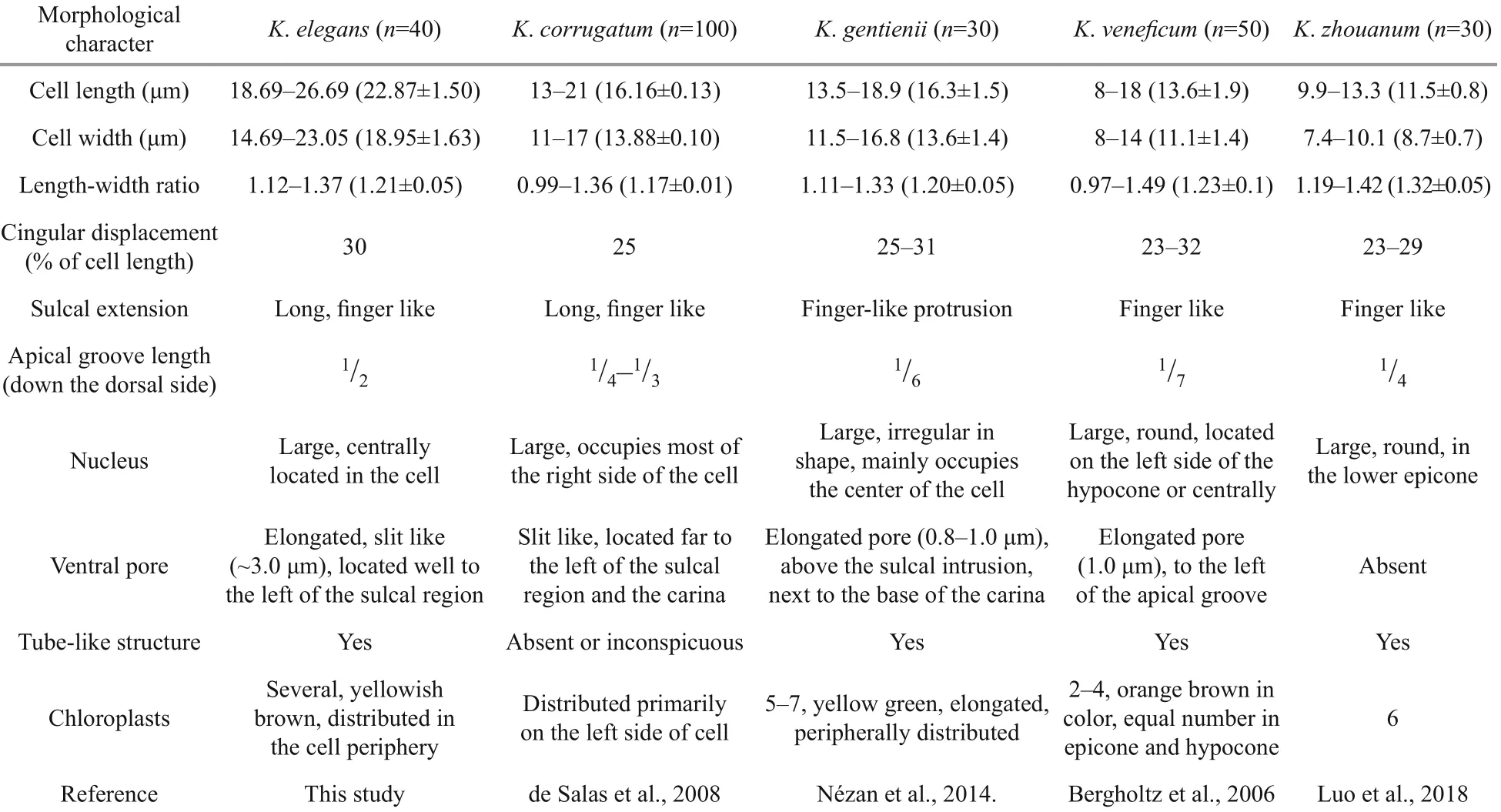
Table 1 Morphological characters of Karlodinium elegans, in comparison with the related Karlodinium species
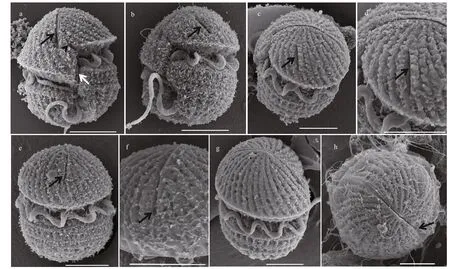
Fig.3 Scanning electron microscopy of Karlodinium elegans strain PTB601 f ixed in O s O 4
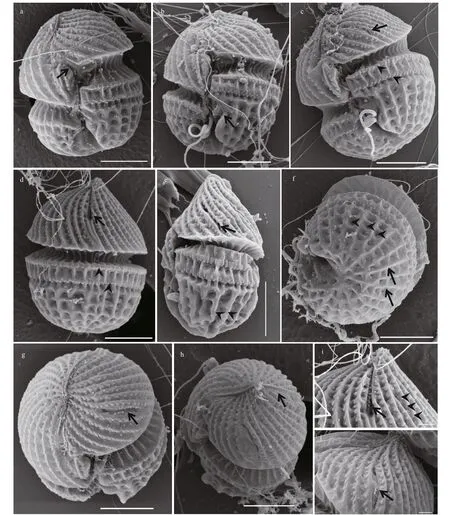
Fig.4 Scanning electron microscopy of Karlodinium elegans strain PTB601 f ixed in a mixture of Lugol’s solution and glutaraldehyde
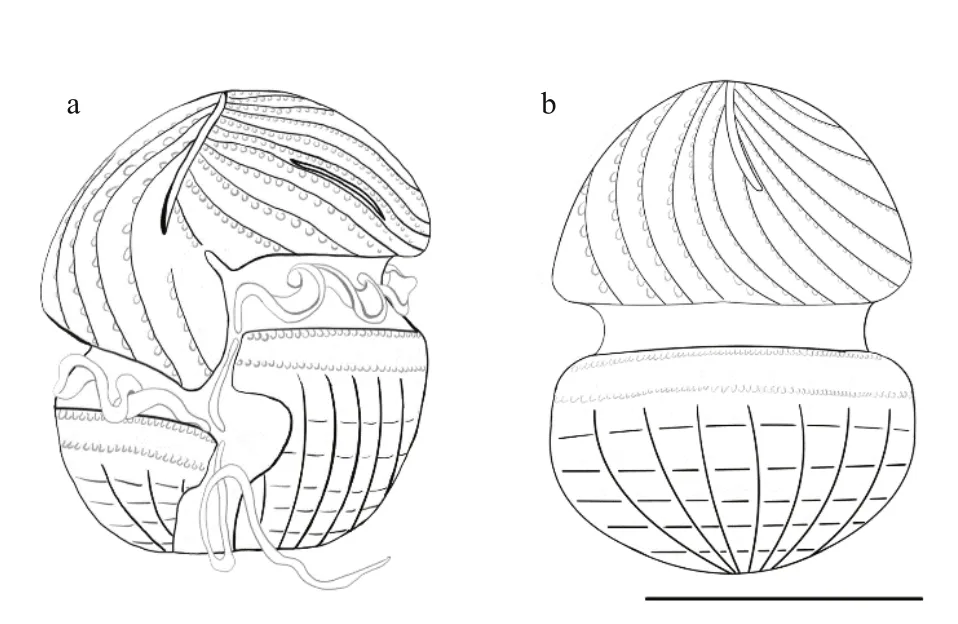
Fig.5 Schematic illustration of Karlodinium elegans sp. nov. based on information obtained from scanning electron microscopy
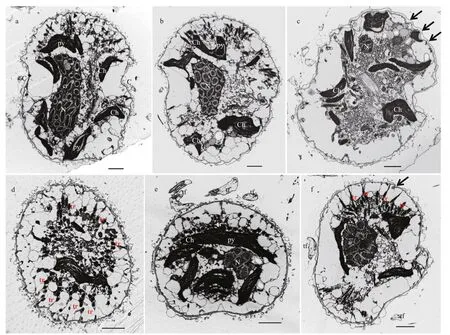
Fig.6 Transmission electron microscopy of Karlodinium elegans strain PTB601

Fig.7 Transmission electron microscopy of Karlodinium elegans strain PTB601
Scanning electron microscopy (SEM)
In the cells fixed in O s O4, the cell surface was covered with a layer of membranous material; however, the SEM still revealed a more detailed morphology than LM. The epicone was conical, and the hypocone was hemispherical (Fig.3a & b). The sulcus intruded into the epicone as a long finger-like structure, and a tube-like structure was present in the intercingular region of the sulcus (Fig.3a & b). In ventral view, the cingulum was seen to be deeply incised and left-handed, with a displacement of about 30% of the cell length; the apical groove was long, extending to the right of the sulcal intrusion (Fig.3a). Many parallel striations were evenly distributed on the surface of the epicone in a left-ward spiral (Fig.3a & c). An elongated slit (hereafter referred as “slit-like ventral pore”) rather than the ventral pore in other Karlodinium species was located on the left side of the epicone and well away from the apical groove (Fig.3a-d). The apical groove was narrow in the middle, wider at the both ends, and extended halfway down the dorsal side of the epicone (Fig.3e-h).
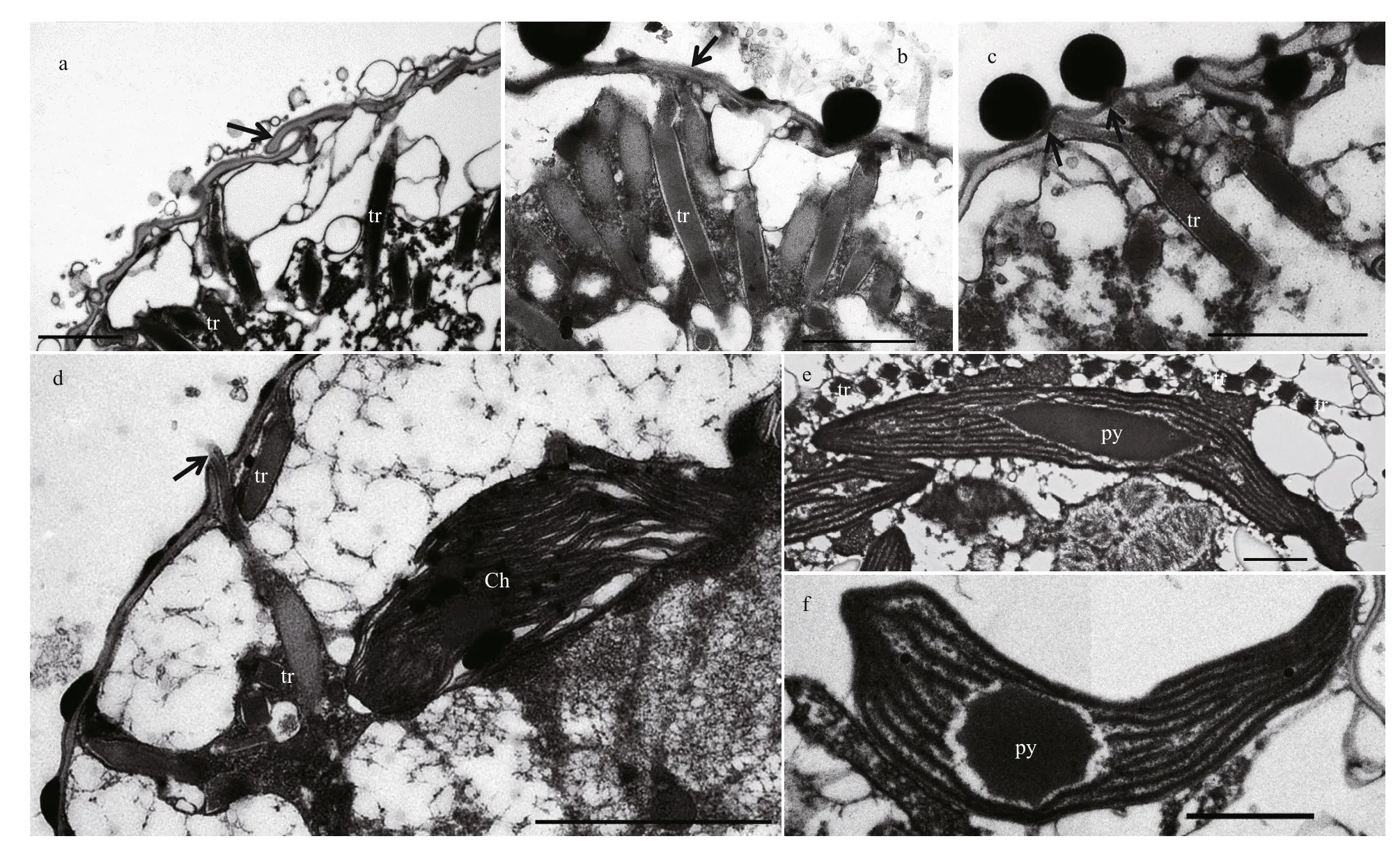
Fig.8 Transmission electron microscopy of Karlodinium elegans
Cells fixed in a mixture of Lugol’s solution and glutaraldehyde preserved the characteristic and distinct details of the amphiesma well. The sulcus was S-shaped, with a curvature on the left lobe of the hypocone near the longitudinal flagellum pore (Figs.4a-c & 5a). The sulcus intruded onto the epicone as a finger-like projection, at ca. 45° to the right (Fig.4a & b). Approximately 32 parallel striations were evenly distributed on the surface of epicone, and curved to the left (Figs.4a-e, 5a & b). A row of amphiesmal knobs (numbering ca. 30 in 10 μm, and 100 nm in diameter) ornamented the right side of each striation (Figs.4a-j, 5a & b)). The elongated and slit-like “ventral pore” was evident, 2.26-3.77 μm in length (Figs.4c, e, j, & 5a). Two horizontal rows of amphiesmal knobs were present on the upper hypocone, the first row covering the lower rim of the cingulum, and the second row was located 1.66-2.00 μm beneath the first row (Figs.4a-d, 5a, & b). The surface of the hypocone was ornamented with the most unusual longitudinal and horizontal stripes (Figs.4a-f, 5a & b). The longitudinal stripes originated radially from the posterior end of the hypocone, the horizontal stripes were disconnected by the longitudinal stripes, and the longitudinal and horizontal stripes formed deep quadrilateral pits, a configuration never reported in the Kareniaceae before (Figs.4a-f, 5a & b). The apical groove was long and straight, starting from the right of the sulcal intrusion on the ventral side of the cell and extending halfway down the dorsal side of the epicone (Figs.4g, h, 5a & b). The apical groove was narrow at the cell apex, but wider at the ends, with two thick rims (Fig.4h & i). Discharged trichocysts are visible in most figures (especially in Fig.4b-d).
Transmission electron microscopy (TEM)
Some details of the ultrastructure of K. elegans observed under the TEM are shown in Figs.6-8. The longitudinal section from the ventral view showed the general arrangement of cell organelles (Fig.6a & b). The dinokaryon contained regularly arranged condensed chromosomes, and was centrally located in the cell, extending from the epicone to the hypocone (Fig.6a & b). The cell membrane was wavy, in accordance with the corrugated cell surface observed both in LM and SEM (Fig.6c). The epicone was dorso-ventrally flattened (Fig.6d), but in a transverse section through the cingulum the cell was seen to be only slightly dorso-ventrally flattened and only the ventral side of the cell was flattened (Fig.6e). Trichocysts were distributed peripherally throughout the cell (Fig.6e), and sometimes densely aggregated in the epicone (Fig.6b & f).

Fig.9 Pigment chromatogram of Karlodinium elegans (PTB601)
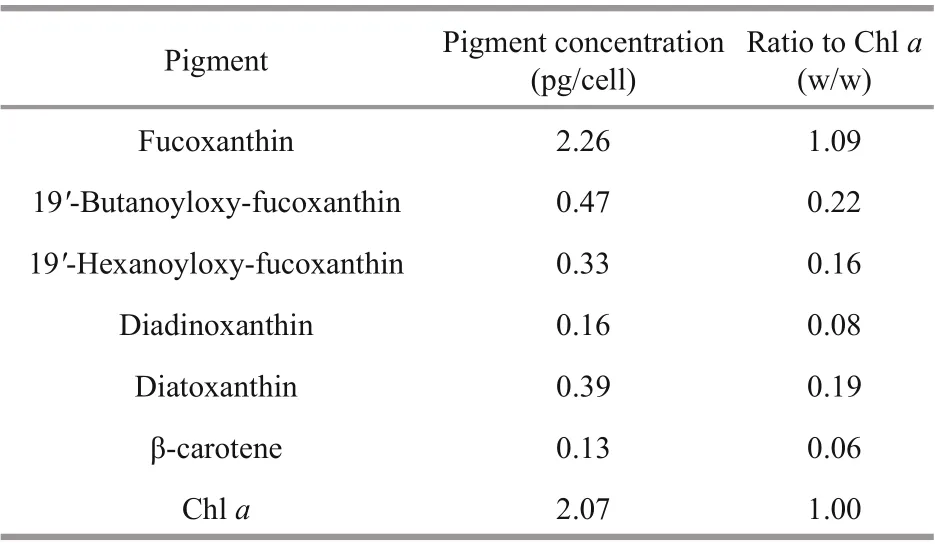
Table 2 Pigment composition and ratios to Chl a of Karlodinium elegans (PTB601)
The cell was well covered by amphiesma, which is well preserved in Fig.7a. A continuous series of amphiesmal vesicles surrounded the cell but no plates or other layers were observed in the vesicles (Fig.7a & b). A relatively thick pellicular layer lay beneath the amphiesmal vesicles; it was wavy, in contrast to what has been observed in other Karlodinium species (Fig.7a & c). In an oblique longitudinal section plane (about 30° left to the longitudinal axis), the slit-like “ventral pore” and finger-like sulcus intrusion were visible; the amphiesma vesicles were clearly present on the surface of the sulcus intrusion, but no vesicles were seen on the slit-like “ventral pore” (Fig.7b). In some cells, a row of small vesicles underlay the inner surface of the pellicle (Fig.7d-f). Many trichocysts were located beneath the pellicular layer, and perpendicular to the cell surface (Fig.8a-c). The mature trichocysts were elongated with a neck, and projected through the pellicular layer when they exocytosed, usually in cells where the amphiesmal vesicles were absent (Fig.8c & d). In cross-sections trichocysts were square (Fig.8e); each chloroplast contained a lenticular and spherical pyrenoid and approximately 10 thylakoid bands (Fig.8d-f).
3.2 Pigment profile
HPLC pigment analyses revealed the pigment profile of K. elegans (PTB601), which included seven main pigments, fucoxanthin, diadinoxanthin, diatoxanthin, chlorophyll a (Chl a), β-carotene, 19′-butanoyloxy-fucoxanthin, and 19′-hexanoyloxyfucoxanthin (Fig.9). Fucoxanthin was the major pigment, Chl a the second one, and no gyroxanthindiester or gyroxanthin-like pigments were detected. Pigment ratios to Chl a based on weight are shown in Table 2.
3.3 Phylogenetic relationships
ITS and LSU rDNA sequences of K. elegans (strains PTB601 and PTB602) were obtained and deposited in GenBank under accession numbers MT161377 (1 016 bp), MT161378 (1 017 bp), MT161379 (668 bp), and MT161380 (638 bp). Partial sequences, 712 bp for LSU rDNA and (609 bp) for ITS, were selected for the phylogenetic tree construction. The two strains of K. elegans had the same LSU rDNA sequences and differed by 2 bp in ITS in the selected region. In the phylogenetic tree based on LSU rDNA, the two K. elegans strains formed a single clade with bootstrap/posterior probability values of 96%/1, and the clade was closest to Karlodinium sp. (strain IFR981, KJ508386 &IFR797, KJ508385) and K. corrugatum (strain KDGSO08, EF469233); the three species clustered in a single branch with bootstrap/posterior probability values of 99%/1 (Fig.10). Karlodinium elegans differed from Karlodinium sp. (strain IFR981 & IFR797, from the French Atlantic and the Mediterranean, respectively) in 6 bp, and from K. corrugatum (from the Southern Ocean) in 34 bp out of the total 712 bp. The genetic distance ( p-distance, pairwise) among Karlodinium species on the basis of LSU sequences (712 bp) is shown in Table 3. The overall variability among species in the genus Karlodinium ranged from 0 to 14.4%; the genetic distance between K. elegans and Karlodinium sp. was 0.6%-0.8%, and 3.3% between K. elegans and K. corrugatum, and the difference to other species was 7.1%-11.9%. Karlodinium elegans and the Karlodinium sp. (strain IFR981 & IFR797) appear to be conspecific.

Table 3 Estimated uncorrected genetic distance ( P-distance, pairwise) among Karlodinium species on the basis of LSU region sequences (712 bp)
The ITS sequences for the French Karlodinium sp. (strains IFR981 & IFR797) and K. corrugatum were unavailable, and K. elegans was most related to K. jejuense, K. zhouanum, and Karlodinium sp. (strain KAMS0708) in the ITS tree, with bootstrap/posterior probability values of 81%/0.94 (Fig.11). The betweenspecies ITS genetic distances ( p-distance, pairwise)in the genus Karlodinium ranged from 0.4% to 25.6% (Table 4). The lowest distances (0.4%) was caused by the high similarity between K. zhouanum and K. jejuense, which were regarded as a same species according to the almost identical rDNA sequences (0-0.2% of LSU rDNA) and only minor morphological differences (Li and Shin, 2018; Luo et al., 2018; Benico et al., 2020). The remaining between-species ITS1 genetic distances were relatively high, ranging from 7.0% to 24.5%. The ITS1 divergence of K. elegans to the other Karlodinium species was 11.4%-25.6%. Additionally, inferred from the phylogenetic tree based on both LSU and ITS sequences, the Karlodinium species clustered into three distinct clades with high support values: Clade 1 consists of K. decipiens and “ K. digitatum” (HK1) ( K. digitatum HK1 may be a misidentification of K. decipiens (Cen et al., 2020), thus, double quotesare used here); Clade 2 consists of K. armiger, K. australe, and K. digitatum; and Clade 3 includes K. veneficum, K. jejuense, K. zhouanum, and K. elegans. It is noteworthy that Clade 1 is much closer to the genus Takayama than to other Karlodinium species in the phylogenetic tree.
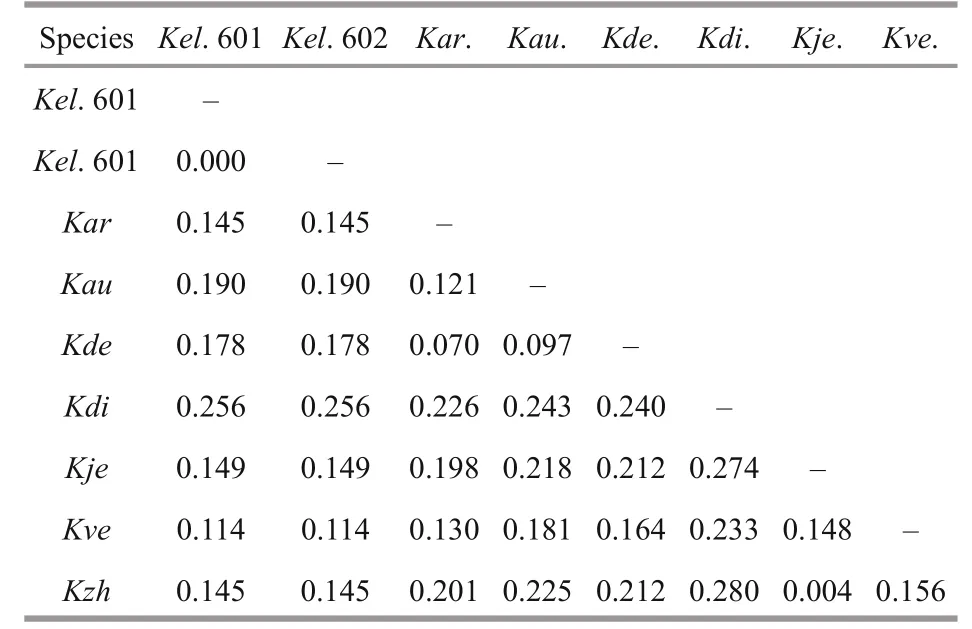
Table 4 Estimated uncorrected genetic distance ( p-distance, pairwise) among Karlodinium species based on partial ITS sequences (569 bp)
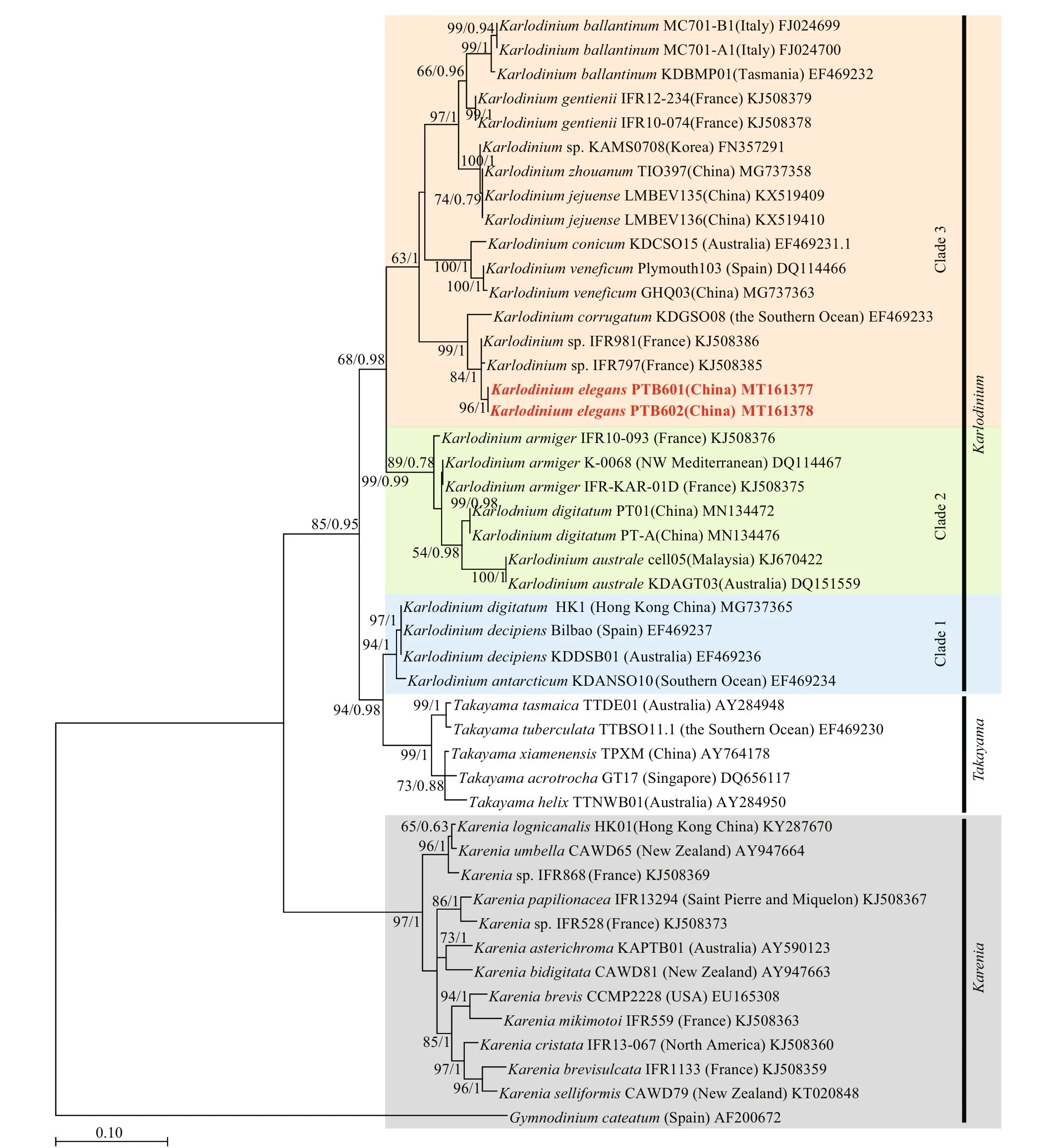
Fig.10 Maximum-likelihood phylogenetic tree of LSU sequences of Karlodinium elegans and closely related species
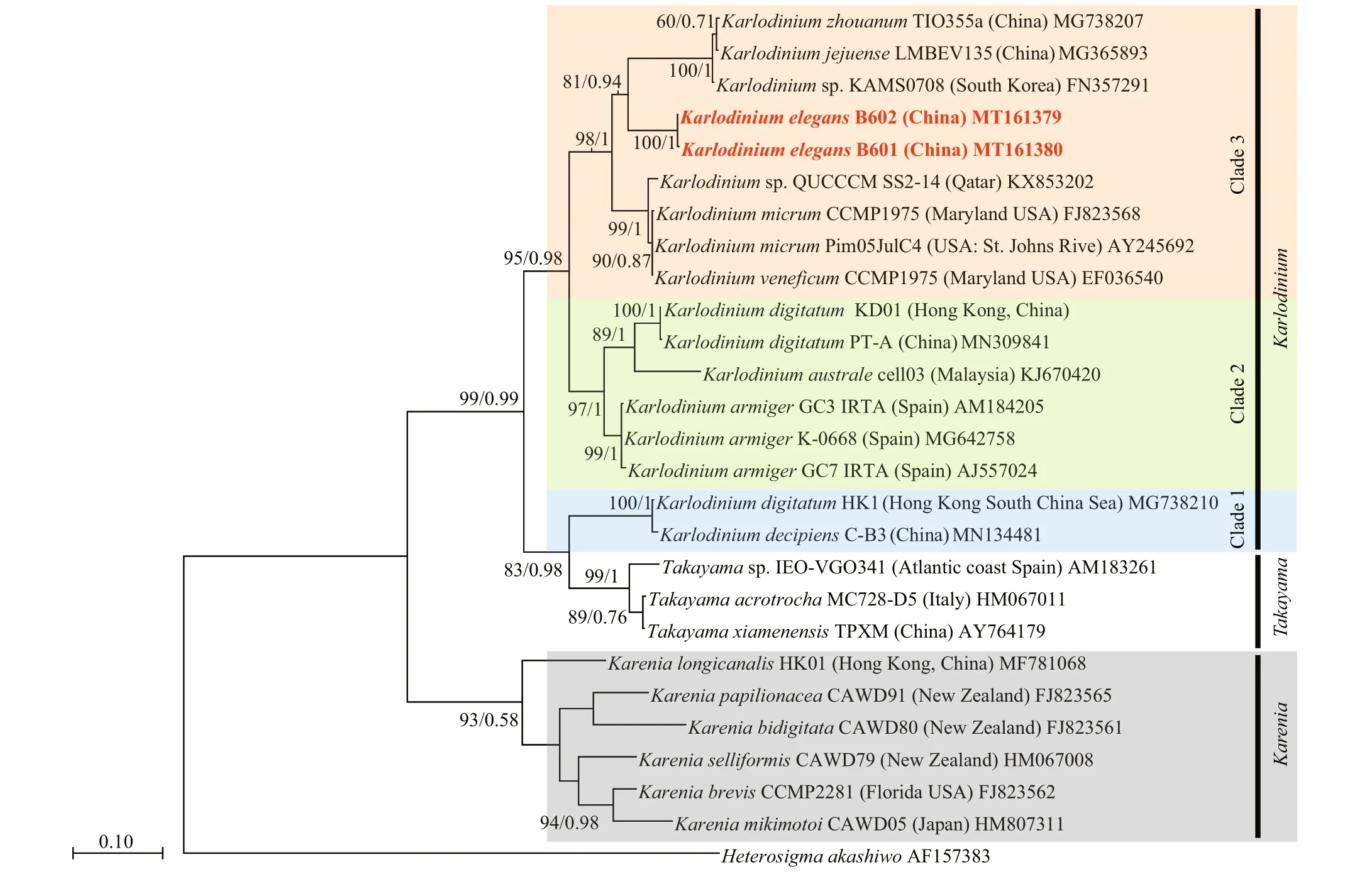
Fig.11 Maximum-likelihood phylogenetic tree of Karlodinium elegans and closely related species inferred from ITS sequences
3.4 Haemolytic activity
The acute toxicity assay of K. elegans (strain PTB601) on A. salina and O. melastigma showed no apparent biological toxicity on the two marine organisms. After a 48-h exposure to K. elegans, no adverse effects nor mortality was observed on either A. salina or O. melastigma.
4 DISCUSSION
On 24-29 May, 2019, a bloom dominated by two species of dinoflagellates, Prorocentrum donghaiense and Karlodinium digitatum occurred in Pingtan coastal areas of Fujian, SE China. The bloom led to massive mortality of cage-cultured fish, which was thought to be caused by K. digitatum (Cen et al., 2020); an unknown dinoflagellate species was also isolated from the seawater samples during the bloom monitoring, and this is described as K. elegans in the present study. Karlodinium elegans is unarmored, the chloroplasts containing internal, lenticular pyrenoids, fucoxanthin as main accessory pigment, and a straight apical groove, all being typical features of the genus Karlodinium. Karlodinium elegans differs from other species of Karlodinium by possessing a long slit-like “ventral pore”, longitudinal striations curving to the left side on the epicone, and a very special and unusual surface ornamentation on the hypocone. Two parallel rows of amphiesmal knobs are present on the hypocone, and the upper row running just below the lower edge of the cingulum, and the lower row locats 1.66-2.00 μm beneath the upper row. The surface of the hypocone is ornamented with the most unusual longitudinal and horizontal stripes. The longitudinal stripes radiate evenly from the antapex of the hypocone and extend to near the lower row of the amphiesmal knobs. The horizontal stripes on the hypocone are parallel and evenly arranged, but disconnected by the longitudinal stripes into short strips, the two sets of stripes together forming quadrangular pits. This type of hypocone ornamentation has not been seen before in the genus Karlodinium.
Morphologically, K. elegans is most similar to K. corrugatum, both having parallel folds on the epicone, which are visible under LM and SEM. Moreover, instead of a pore-like ventral pore, a long slit-like “ventral pore” is located on the left of the sulcus intrusion in both K. corrugatum and K. elegans. The epicone folds on the two species are different. In K. corrugatum the folds are parallel and extend radially from the cell apex (Fig.11d in de Salas et al., 2008), while in K. elegans, the folds are parallel and spiral to the left. No ornamentation on the hypocone is mentioned in K. corrugatum (de Salas et al., 2008). Karlodinium corrugatum also differs from K. elegans in the nucleus occupying most of the right side of the cell (Figs.7d & 11d in de Salas et al., 2008), while the nucleus in K. elegans is centrally located. A final difference between the two species is observed in the apical groove, which in K. corrugatum extends1/4-1/3down the dorsal side of the epicone (de Salas et al., 2008), while in K. elegans it extends halfway down the epicone.
Among other species of Karlodinium, K. gentienii also has parallel corrugations, rows of amphiesmal knobs on the epicone, and two lines of knobs on the hypocone, which are very similar to those of K. elegans. Karlodinium gentienii is smaller than K. elegans, however, with an average cell size of 18.9 μm in length and 13.5 μm in width, comparing to 22.87 μm and 18.95 μm, respectively, in K. elegans. Karlodinium gentienii further differs from K. elegans in having a short (less than 1 μm) ventral pore located above the sulcal intrusion and close to the base of the apical groove (Fig.6a & b in Nézan et al., 2014), comparing to the ca. 3-μm long slit in K. elegans, which is located far from the apical groove.
The presence of a ventral pore in Karlodinium is one of the key morphological feature thought to distinguish Karlodinium from Karenia (Daugbjerg et al., 2000; Bergholtz et al., 2006). However, several new species without a ventral pore have been identified, e.g. K. antarcticum, K. ballantinum, K. zhouanum, K. digitatum, and K. jejuense (de Salas et al., 2008; Li and Shin, 2018; Luo et al., 2018). Bergholtz et al. (2006) observed the ventral pore in K. veneficum to be covered by amphiesmal cisternae on the outside and internally by a flattened vesicle; however, in K. elegans no vesicle was observed in the slit pore.
In SEM preparations of Karlodinium, OsO4is the most commonly used fixative (Bergholtz et al., 2006; de Salas et al., 2008; Li and Shin, 2018; Luo et al., 2018). In the study of K. gentienii by Nézan et al. (2014), a double fixation (Lugol’s solution and glutaraldehyde) was used and compared to a fixation in only OsO4. More details of the cell surface were obtained with the double fixation. We used both fixation schedules in K. elegans, and both displayed similar morphological details. However, the double fixation revealed a much clearer cell surface. Therefore, the use of double fixation appears to be preferable for SEM preparations of Karlodinium species.
The family Kareniaceae is characterized by haptophyte types of pigments (Jeffrey et al., 2011), and the chloroplasts contain fucoxanthin or fucoxanthin-derivatives but no peridinin (Bergholtz et al., 2006). According to the six pigment-based dinoflagellate chloroplast types proposed by Zapata et al. (2012), species in the genus Karenia grouped in the Chloroplast Type 2, with Fuco, 19’-acyloxyfucoxanthins and their keto derivatives, and gyroxanthin diesters as signature pigments; species in the genus Karlodinium clustered in Chloroplast Type 3, which resembled Type 2 but lacked the fucoxanthin of Karenia species, while Butfuco and Hex-fuco were the major acyloxyderivatives; species of Takayama also grouped into Type 3, but lacked gyroxanthin diesters. Pigment fingerprints of K. elegans were in agreement with Type 3, with fucoxanthin as the major carotenoid, But-fuco and Hex-fuco the major acyloxyderivatives, and no keto-19’-acyloxyderivatives. The gyroxanthin diesters of Karlodinium and Karenia species were not detected in K. elegans. Pigment profiles of 7 species of Karlodinium, K. armiger (Bergholtz et al., 2006; Garcés et al., 2006; Zapata et al., 2012), K. australe (de Salas et al., 2005), K. veneficum (Garcés et al., 2006; Bachvaroff et al., 2009; Zapata et al., 2012), K. decipiens (Laza-Martinez et al., 2007; Zapata et al., 2012), K. gentienii (Nézan et al., 2014), K. zhouanum (Luo et al., 2018) and K. elegans (present study) are presently available; four of the 7 species, K. australe, K. gentienii, K. zhouanum and K. elegans lack gyroxanthin diester, indicating that the gyroxanthin diester is not a genetic marker of the genus Karlodinium .
The between-species ITS1 genetic distances in the genus Karlodinium ranged 7.0%-24.5%. The ITS1 divergence of K. elegans to the other Karlodinium species was 11.4%-25.6%, and was much higher than 4%, which is a distance used to delineate most freeliving dinoflagellate species (Litaker et al., 2007), suggesting that K. elegans is genetic separated from the ITS sequences available Karlodinium species. In the molecular phylogenetic tree inferred from LSU rDNA, K. elegans formed a single clade with K. corrugatum (EF469233) and Karlodinium sp. (KJ508385, KJ508386), with a high support value (99%/1). As reported by Nézan et al. (2014), the interspecific LSU rDNA variability within the genus Karlodinium ranged 2.0%-9.9%, and the intraspecific variability ranged 0-1.5%. K. elegans differed from K. corrugatum (from the Southern Ocean) at 34 loci in the LSU rDNA region (712 bp), with a genetic distance of 3.3%; the distance exceeds the maximum value of the intraspecific divergence between individuals of a same species, and is higher than the minimum distance of the interspecific divergence within the genus Karlodinium, supporting the notion that K. elegans is separate from K. corrugatum. Karlodinium elegans was, however, closely related to two Karlodinium sp. strains known as IFR981 and IFR797 from France (Atlantic coast and Mediterranean coast, in Nézan et al. (2014)), with a genetic distance of 0.6% and 0.8%, respectively. The distance between K. elegans and Karlodinium sp. is at the intra-species level, and the two species could not be resolved in the LSU-based phylogeny. The two strains of Karlodinium sp. most likely belong to K. elegans, however, this needs to be confirmed by morphological studies.
Based on both LSU sequences and ITS sequences in the present study, the genus Karlodinium is paraphyletic and clusters into three clades. Some species, e.g. K. decipiens and K. antarcticum are genetically more related to Takayama spp. suggesting they may have a common ancestor with Takayama; this agrees with de Salas et al. (2008) and Siano et al. (2009). However, there is still no morphological character to distinguish the three clades. The discovery of more novel species or more detailed ultrastructure features may help to clarify the taxonomy of the two genera Karlodinium and Takayama.
The toxicity test revealed that the culture of K. elegans has no acute toxic effect on brine shrimp and marine medaka in laboratory. Some toxic Karlodinium species were reported to lose toxicity after being cultured in laboratory for some time (Yang et al., 2019). However, in our case, the K. elegans culture used was newly established. This indicates that K. elegans is a non-toxic species or the toxicity was below the level of detection. Future studies on toxicity should involve grazers as the presence of grazers or competing species can induce toxicity production of some phytoplankton (Tammilehto et al., 2015; Chakraborty et al., 2016).
5 CONCLUSION
A naked dinoflagellate was isolated from a bloom in May 2019 in Fujian coastal waters, SE China. The species was identified as a new species in genus Karlodinium: K. elegans sp. nov. based on morphological, molecular, and pigment composition analyses, as well as comparison with closely related species of the genus. The acute toxicity tests revealed no acute toxic effect of K. elegans on brine shrimp and marine medaka in the laboratory. It is noteworthy as a final note that the species diversity in the Kareniaceae is underestimated, and more Kareniaceae species were co-occurred in the Karenia bloom in Chinese coastal waters.
6 DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
7 ACKNOWLEDGMENT
Special appreciations are expressed to Prof. Moestrup of the University of Copenhagen for his helpful comments and for improving the English text. We sincerely thank Dr. Tao JIANG of the Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences for his help in the pigment composition analysis. We deeply thank Prof. Hallegraeff of University of Tasmania for his kind help on the new species confirmation.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Influence of sequential tropical cyclones on phytoplankton blooms in the northwestern South China Sea*
- Simulated perturbation in the sea-to-air flux of dimethylsulfide and the impact on polar climate
- Performance of ecological restoration in an impaired coral reef in the Wuzhizhou Island, Sanya, China*
- Investigating factors driving phytoplankton growth and grazing loss rates in waters around Peninsular Malaysia
- Effects of oxytetracycline dihydrate and sulfamethoxazole on Microcystis aeruginosa and Chlamydomonas microsphaera*
- Reproductive cycle of Ophiopholis mirabilis (Echinodermata: Ophiuroidea) in Zhangzi Island area, northern Yellow Sea*
