Novel diagnostic approaches for Fanconi anemia(FA)by single-cell sequencing and capillary nano-immunoassay
2021-01-27LixinChngXingjieGoGungzhenJiXuelinChengYoZouToChengWeipingYunXiofnZhu
Lixin Chng,Xingjie Go,Gungzhen Ji,Xuelin Cheng,Yo Zou,To Cheng,Weiping Yun,∗,Xiofn Zhu,∗
aState Key Laboratory of Experimental Hematology,National Clinical Research Center for Blood Diseases,Institute of Hematology& Blood Diseases Hospital,Chinese Academy of Medical Sciences & Peking Union Medical College,Tianjin 300020,China;
bDepartment of Biochemistry and Molecular Biology,School of Basic Medical Sciences,Tianjin Medical University,Tianjin 300070,China
Abstract Next-generation sequencing technology has been widely utilized for the diagnosis of Fanconi anemia(FA).However,mixed cell sequencing and chimerism of FA patients may lead to unconfirmed genetic subtypes.Herein,we introduced two novel diagnostic methods,including single-cell sequencing and capillary nano-immunoassay.One FA case with FANCM c.4931G>A p.R1644Q and FANCD1 c.6325G>A p.V2109I was studied.The DNA of 28 cells was amplified and eight types of cells were observed after Sanger sequencing.There were two homozygous mutations(FANCM/FANCD1).Furthermore,the capillary nano-immunoassay was conducted to analyze the expression profile of FA-associated proteins.Abnormal FANCM and FANCD1 expressions simultaneously existed.This case was thus diagnosed as FA-D1/FA-M dual subtype.Compared with mixed cell sequencing,single-cell sequencing data shows more accuracy for the FA subtype evaluation,while the capillary nano-immunoassay is a good method to detect the expression profile of abnormal or modified FA protein.
Keywords:Capillary nano-immunoassay,FANCD1,FANCD2,FANCM,Fanconi anemia,Single-cell sequencing
1.INTRODUCTION
Fanconi anemia(FA),a genetically heterogeneous rare autosomal recessive or X-linked genetic disorder,is characterized by congenital malformations,hematological problems,and predispositiontomalignancies.1-3Fanconianemiasignalingisreportedly implicated in a series of molecular and cellular processes,such as DNA damage response(DDR),DNA replication,cell cycle,and maintenance of genome stability.4-6To date,22 FA genes have been identified in the FA pathway and all were involved in maintaining genome stability.7-11
According to the different mutated genes in the clinic,Fanconi anemia was divided into 22 genotypes.Different genotypes have a different clinical prognosis,even heterozygous mutations of the FA gene may also have different clinical manifestations.For instance,individuals containing BRCA2 mutations are susceptible to breast cancer.12Heterozygote FANCD2 mutations are linked to the presence of childhood T-Cell acute lymphoblastic leukemia and testicular seminoma.13Hence,accurate genotyping of patients with Fanconi anemia is very important.The development of second-generation sequencing technology provides a good diagnostic method for FA genotyping,but unexplained results still appear in clinical practice.For instance,if a patient carries a heterozygous mutation of both FA genes or a hybrid heterozygous mutation of both FA genes,how to genotyping these patients accurately becomes a clinical problem.
In the present study,we recommended two novel diagnostic approaches for FA,namely single-cell sequencing and capillary nano-immunoassay.We analyzed a FA patient with positive results of comets and chromosome breakage test,significant hematological abnormalities,and susceptible family history,but with the heterozygous mutations of two FANC genes(data of mixed cells sequencing).
2.RESULTS
2.1.Characteristics of FA patient
In this study,the comprehensive clinical manifestations,family history,and laboratory results contributed to the diagnosis of FA for the patients,according to FA diagnostic criteria.2,3,14To further analyze the subtype of FA,we designed a targeted capture sequencing assay to detect 417 blood disease genes,including the seventeen known FA-related genes.As shown in Table 1,we observed the mutations of FANCM c.4931G>A and FANCD1 c.6325G>A(previously reported in breast cancer patients)15for this FA patient and his father.However,there was no mutation for his mother.Besides,his grandmother carried the FANCD1 c.6325G>A mutation,while his grandfather harbored the FANCM c.4931G>A mutation.Figure 1 showed the pedigree chart of this patient family,targeting the above detectable mutations.
The results of the multiplex ligation-dependent probe amplification assay(Fig.2)indicated that there was no deletion or duplication of the given chromosomal regions in both case and normal control.According to the above data,we failed to confirm the FA subtype of this patient.

Table 1 The data of targeted capture sequencing assay.
2.2.Data for single-cell DNA amplification and Sanger sequencing
Chimerism may exist in the FA patient.Therefore,the assay of single-cell DNA amplification and Sanger sequencing was carried out.As shown in Table 2,twenty-eight cells were successfully amplified,and eight types of cells were observed.The peak map data of Sanger sequencing for eight cell types were shown in Figure 3.There were two homozygous mutations(FANCM c.4931G>A and FANCD1 c.6325G>A)in this case.Interestingly,the rate of cells with at least one homozygous mutation was 42.9%(12/28),which is similar to the rate of comet cells(39%).
2.3.Analysis of FA protein expression by capillary nanoimmunoassay
Functional experiments of pathogenic mutations are still required after the analysis of single-cell sequencing.Here,the capillary nano-immunoassay via NanoPro 1000 system was performed to detect the expression profile of three FA-associated proteins,including FANCD2,FANCM,and FANCD1.GAPDH protein was used as a control(Fig.4A).The expression level of the targeting protein should be normalized to the GAPDH control.The disturbance of FANCD2 mono-ubiquitination in response to MMC is considered as a key feature of FA patients.16,17As shown in Figure 4B,the treatment of MMC led to the presence of a strong peak signal(pI=5.19)of FANCD2 protein in normal control(NC),but not positive control(PC)or this case.The expression map mode of FANCD2(Fig.4C)showed that a new band(pI=5.19)appeared under the treatment of MMC.This strong peak signal is more likely to come from the MMC-induced FANCD2 mono-ubiquitination.In addition,the MMC-induced expression differences were observed for FANCM(Fig.4D)and FANCD1(Fig.4E)in normal control,rather than the case.The column chart of protein expression(Fig.4F)further showed that the exposure of MMC affects very little of the normalized chemiluminescence value of FANCD2,FANCM,and FANCD1 in case,while the impact on normal control is very prominent.The phenomenon indicated the protein abnormality of both FANCM and FANCD1 in the patient.This case was thus diagnosed as FA-D1/FA-M dual subtype.
3.DISCUSSION
The diagnosis of FA subtype depends on the existence of homozygous,compound heterozygous,biallelic,or dominantnegative mutation for one FANC gene.16,18,19However,our previous data of whole-exome sequencing showed that there are multiple heterozygous FANC gene mutations in the same FA patient.20These mutant modes could not be interpreted by the classic FA genetic approach,which brings confusion to the diagnosisof the FA subtype.The heteroskedasticof cellsamples for next-generation sequencing and chimerism of FA patients should be considered.
Although traditional complementation analyses can provide more accurate analysis,21the time-consuming and complicated procedures put a limit on the wild application in molecular genetic testing laboratories.To address this issue,two novel diagnostic approaches for FA,namely single-cell sequencing and capillary nano-immunoassay,were utilized.
The current second-generation sequencing based on peripheral blood mixed cells is not feasible for the accurate classification of certain FA patients.Single-cell DNA amplification and Sanger sequencing can detect the specific mutation type,chimeric state,or subtype of FA patient,which makes up for the false-negative results of previously mixed cell sequencing to a certain degree.Additionally,capillarynano-immunoassay,basedontheisoelectric point(pI)of protein,requires very small amounts of cell samples and provides the information of different isomers,which exhibits higher sensitivity than traditional western blotting assay.22,23BecausepIvalue issensitive todifferentisomers,themutantorposttranscriptional modified proteins exhibit the different peak signal,and the size of the area under the curve presents the expression levels of protein.
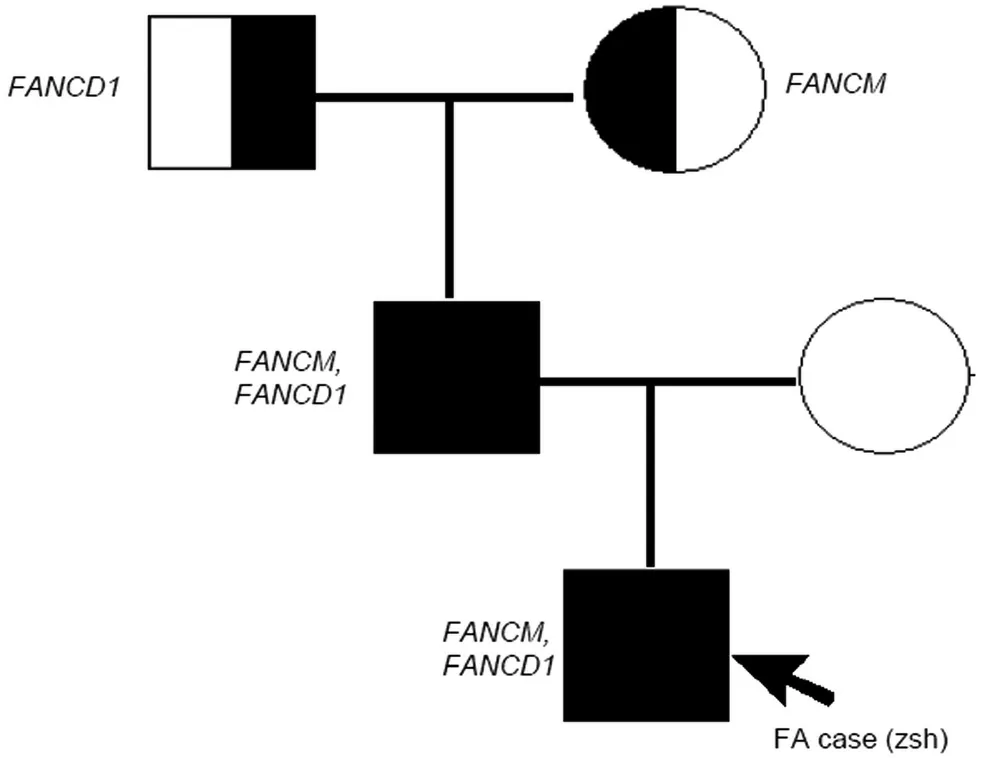
Figure 1.The pedigree chart of FA patient family,targeting the heterozygous mutation of FANCD1 and FANCM.FANCM:FANCM chr14-45658156 c.4931G>A mutation; FANCD1:FANCD1 chr13-32914817 c.6325G>A mutation.
Here,FA-D1/FA-M dual subtype was observed in this case,by the ways of single-cell sequencing and capillary nanoimmunoassay.The complexity and variability of cell division and proliferation may result in the simultaneous occurrence of two or more FA patterns in one patient,and the incidence of FA is likely to depend on the proportions of the cell types.

Figure 2.The data of MLPA assay using four probes.(A)P031-B1/P032-B1 FANCA;(B)P113-A2 FANCB;(C)p260-B1 PALB2-RAD50-RAD51C-RAD51D;(D);P057-B1 FANCD2.

Table 2 The result of single cell DNA amplification and Sanger sequencing.
In summary,the combination of single-cell sequencing and capillary nano-immunoassay favors the diagnosis of FA subtype,which provides a new diagnostic method targeting the confusing results or unconfirmed genetic subtype,such as the heterozygous mutations of two FA genes in one FA patient.

Figure 3.The peak map data of single cell DNA amplification and Sanger sequencing assay.
4.MATERIAL AND METHODS
4.1.Patient
An eight-year-old boy was enrolled in our hospital due to the pancytopenia for one mouth in 2008.The patient had severe anemia on presentation,appeared petechia on the skin,without any malformation.Routine blood test data:white blood cell(4.27×109/L),redbloodcell(1.83×1012/L),hemoglobin(65g/L),platelet(16×109/L).Marrow film data:hyperplasia.Chromosome:“46,XY[20]”.The results of Mitomycin C(MMC)induced chromosome fragility test showed that the rate of chromosomal abnormality cell is 29%(>15%is considered as a positive result),while the data of single-cell gel electrophoresis test indicated that comet cell rate equals to 39%(>30%is considered as a positive result).Additionally,his father has thrombocytopenia,the rate of chromosomalabnormalitycellis16%,andcomet cellratesis39%;however,his mother,grandmother,and grandfather are healthy.
4.2.Targeted capture sequencing assay
A total of 417 blood disease genes were enriched through a biotinylated capture probe(MyGenostics,Baltimore,MD,USA),as described previously.24Sanger sequencing was then performed to analyze the potential gene mutations.The primers used for Sanger sequencing were shown in Table 3.
4.3.Multiplex ligation-dependent probe amplification(MLPA)
Salsa MLPA Probemix Kit was used to detect the potential mutation sites,based on the probes of P031-B1/P032-B1 FANCA,P113-A2 FANCB,p260-B1 PALB2-RAD50-RAD51C-RAD51D,and P057-B1 FANCD2.One healthy individual was included as normal control.
4.4.Single-cell DNA amplification and Sanger sequencing
Fluorescence-Activated Cell Sorting system was used to directly sort 50 peripheral blood lymphocytes of the patient into 50 wells of 96-well plate.A single-cell DNA amplification kit(MALBAC TM Single-cell WGA Kit)was then used to extract and amplify the DNA.Finally,FANCM chr14-45658156 c.4931G>A and FANCD1 chr13-32914817 c.6325G>A mutations of each sample were detected by Sanger sequencing.
4.5.Capillary nano-immunoassay
Atleast 2000peripheral bloodlymphocytes were culturedina 6-well plate for 18hours,in the presence or absence of 300ng/mL MMC.The cell lysate was harvested in the ice-cold Bicine/CHAPS Lysis Buffer(ProteinSimple,Santa Clara,CA)plus DMSO Inhibitor Mix and Aqueous Inhibitor Mix(Protein Simple)in ice condition for 30min.Then,6μL Premix G2(pH 3-10)plus isoelectric point(pI)Standard Ladder 1(Protein Simple)were mixed with the lysate in 3:1.The primary antibodies(1:50 dilution),including rabbit anti-FANCD2(ab108928,Abcam),rabbit anti-FANCD1(ab27976,Abcam),rabbit anti-FANCM(ab95014,Abcam),and rabbit anti-GAPDH(#5174,Cell Signaling Technology),were then added.GAPDH was used as an internal control.Next,an anti-human IgG-HRP secondary antibody(Protein Simple)was diluted 1:100 into antibody dilution buffer and mixed with luminol/peroxide at a 1:1 ratio.After the additionofsecondaryantibody andluminol/peroxide,eachsample was loaded on the NanoPro 1000 system.Emitted light was quantified.The compass software 2.5.11 was utilized to identify and quantify the chemiluminescent peaks and visually optimize tracings.Besides,one healthy individual was included as a normal control; and another patient with compound heterozygous mutation of FANCA(FANCA c.3638_3639del p.1213_1213del het and FANCA c.3348+1G>A splicing het)was also enrolled as the PC of Capillary nano-immunoassay.

Figure 4.The expression profiles of FANCD2,FANCM and FANCD1 proteins via capillary nano-immunoassay.(A-E)The expression profiles of FA-associated proteins,including FANCD2,FANCM and FANCD1.Cells were untreated(-)or treated(+)with 300ng/mL MMC.GAPDH protein was used as a control.The expression map mode of FANCD2 was shown in(C);(F)The column chart of protein expression in the cells of normal control and case.The chemiluminescence value of FA-associated proteins was normalized to that of GAPDH control protein.NC = normal control,PC = positive control,pI = isoelectric point.

Table 3 The primer sequences for the targeted capture sequencing assay.
ACKNOWLEDGMENTS
This work was partially supported by grants from the National Natural Science Foundation of China(81670112 and 31571380); CAMS Innovation Fund for Medical Sciences(CIFMS,2016-I2M-1-002); The National Key Research and Development Program of China(2016YFC0901503).We thank Li Zhang,Congcong Sun,and Chao Liu(Institute of Hematology & Blood Diseases Hospital)for their excellent technical assistance and helpful comments.