CRISPR/Cas9-based Editing of Acetyl-CoA Carboxylase (ACC1) Gene in Barley
2021-01-15LinMengmengSunMengChenFengjuanLyuBoNiFeiWuJiajieAllanCaplanandFuDaolin
Lin Meng-meng , Sun Meng Chen Feng-juan Lyu Bo , Ni Fei Wu Jia-jie Allan Caplan, and Fu Dao-lin,
1 State Key Laboratory of Crop Biology, Shandong Agricultural University, Taian 271018, Shandong, China
2 Department of Plant Sciences, University of Idaho, Moscow, ID 83844, USA
3 Center for Reproductive Biology, Washington State University, Pullman, WA 99164, USA
Abstract: Plastid localized acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) is a target for aryloxyphenoxypropionates (APPs) and cyclohexanediones (CHDs), two groups of selective herbicides used in controlling grassy weeds. Wheat and barley are important cereal crops in the grass (Poaceae or Gramineae) family, and thus sensitive for those herbicides. Characterization of this form of ACCase (or ACC1) in wheat and barley is essential if these agents are used in the sustainable agriculture. In this study, it was confirmed that a single ACC1 gene presented on the second chromosome per homologous group in common wheat, wild emmer wheat, goat grass and barley. Using CRISPR/Cas9, the barley ACC1 gene was edited, specifically in the carboxyl transferase (CT)domain that was critical for herbicide responses in grass species. Two new alleles were generated, one with a 3-bp deletion leading to ACC1:p.Ile1878del and one with a 26-bp deletion causing ACC1:p.Ser2099_Lys2311del. Both were recovered as heterozygotes in the T0 generation. All the seven T0 plants harboring the 3-bp deletion grew normally, but the only T0 plant with 26-bp deletion died at the extension stage (Zadoks 32), probably because there was inadequate ACC1 activity when the plant was big. In the T1 generation, the 3-bp deletion (or Ile1878del) did not impact the edited plants in tiller numbers, tiller height, spike length and spikelet numbers, when compared to the wild-type allele in the non-edited segregants. This study demonstrated that CRISPR/Cas9 was practical to generate single amino acid deletions in the ACC1 protein and the Ile1878 deletion did not compromise plant growth. Unfortunately, the ACC1:p.Ile1878del protein did not confer resistance to the currently tested APP herbicides, including clethodim, haloxyfop, quizalofop-P-ethyl and sethoxydim.
Key words: ACCase, genome editing, herbicide resistance, Triticeae, wheat
Introduction
A series of innovations over the last 10 years have made genome editing in plants wholly practical.Technologies have advanced from the early Zincfinger nucleases (ZFNs) (Milleret al., 2007), to activator-like effector nucleases (TALENs) (Woodet al., 2011), and finally to today's most used approachthe clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein 9(Cas9) (Doudna and Charpentier, 2014). Genome editing is based on DNA double-strand breaks (DSBs)and the associated repair mechanisms: nonhomologous end-joining (NHEJ) and homology-directed repair(HDR) (Symington and Gautier, 2011). NHEJ can be error prone, introducing small insertions or deletions(InDel) when the DNA ends are rejoined (Gorbunova and Levy, 1997). InDels occurred in a coding region or in a splicing site may disrupt gene function. HDR requires a homologous DNA as a template to assist the DSB repair, and an engineered DNA homolog can be used to incorporate desirable bases to a target gene (Huang and Puchta, 2019). Even more selective changes can be made in plants using a combination nickase and deaminase (Zonget al., 2017) and other CRISPR-based approaches (Hahnet al., 2018; Huang and Puchta, 2019).
In prokaryotes, CRISPR acts as an immune system to defeat invading viruses (Barrangouet al., 2007;Sampsonet al., 2013). There are six types of CRISPR systems (I-VI) (Wiedenheftet al., 2012; Sinkunaset al., 2013; Niewoehneret al., 2014; Terns, 2018).The CRISPR/Cas9, a type II system (Terns, 2018),requires the Cas9 endonuclease and a single guide RNA (sgRNA) that has a 5' 20-bp target sequence next to a 3' protospacer-adjacent motif (PAM=NGG). Cas9 cleaves the host DNA that is complementary to the 20-bp guide sequence in a sgRNA. To date, CRISPR/Cas9 has successfully edited genomes inArabidopsis(Fauseret al., 2014), tobacco (Gaoet al., 2015), rice(Miaoet al., 2013; Zhouet al., 2014), wheat (Upadhyayet al., 2013) and many other plant species (Sodaet al.,2018).
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2)is a biotinylated enzyme that first acquires the carboxyl group by its biotin-carboxylase activity, and then transfers that to acetyl-CoA using its carboxyl transferase (CT) activity (Nikolauet al., 2003). Most plants have two structurally distinct ACCases, with the heteromeric form in plastids and the homomeric form in the cytosol (Sasaki and Nagano, 2004). The plastid form, which accounts for more than 80% of the total ACCase activity (Albanet al., 1994; Herbertet al., 1996), is responsible for the primary fatty acid biosynthesis (Nikolauet al., 2003), while the cytosolic form is responsible for synthesis of long-chain fatty acids, flavonoids and for malonylation (Post-Beittenmiller, 1996; Sasakiet al., 1995). In eudicot species, such asArabidopsis(Feria Bourrellieret al.,2010) and pea (Sasakiet al., 1993), the plastid ACCase is a heteromeric form. In the grass family (Poaceae or Gramineae), such as in wheat and in rice, only the homomeric forms occur in both cytosol and plastids(Konishi and Sasaki, 1994; Konishiet al., 1996), and both cytosolic and plastid ACCases are encoded by nuclear genes (Gornickiet al., 1994; Egliet al., 1995;Podkowinskiet al., 1996).
The plastid ACCase is inhibited by two chemical classes of herbicides, aryloxyphenoxypropionates(APPs) and cyclohexanediones (CHDs), which target the CT activity, thus blocking the transfer of the carboxyl group to acetyl-CoA (Burtonet al., 1991;Rendinaet al., 1990). The heteromeric plastidlocalized ACCase is insensitive to these herbicides(Albanet al., 1994; Sasaki and Nagano, 2004).However, some grasses, such as wheat and rice, only have the homomeric form in plastids, which explains the origin of herbicide sensitivity in this family. In wheat, point mutations of nuclear ACCase genes have been successfully used to create resistance to an APP-type herbicide, quizalofop (Ostlieet al., 2015).
Using CRISPR/Cas9, engineered herbicide resistance has been achieved by editing the riceEPSPSgene (Liet al., 2016), the riceALSgene (Sunet al.,2016) and the watermelonALSgene (Tianet al.,2018). Barley is an important cereal crop in the world.As a model crop in Triticeae, the whole genome sequence of barley is available (IBSC, 2012; Mascheret al., 2017). Some barley genotypes, such as 'Golden Promise', are amenable for gene transformation,both for biolistic bombardment andAgrobacteriummediated transfer (Lvet al., 2015). Thus far, a number of barley genes have been edited using CRISPR/Cas9(Holmeet al., 2017; Kapusiet al., 2017; Gaspariset al., 2018; Henselet al., 2018; Kapusi and Stöger,2018). In this study, CRISPR/Cas9 was used to edit the barleyACC1 gene, an ortholog of the wheatACC1 gene that conferred resistance to herbicide, quizalofop(Ostlieet al., 2015).
Materials and Methods
Plant materials and growth conditions
Barley variety 'Golden Promise' (Hordeum vulgareL.)was used in this study. Non-transgenic wild-type plants were grown on the research farm of the Shandong Agricultural University, Taian, China and were used to collect immature embryos for genetic transformation.Transgenic plants were grown in a greenhouse that maintained a 16 h photoperiod with day temperature of 22℃-25℃ and night temperature of 15℃-20℃.
CRISPR/Cas9 system
The CRISPR/Cas9 system was developed in two steps.The barleyACC1 sequence (GenBank: MK481066 and MK481069) was firstly used for guiding RNA design. Using CRISPRdirect (http://crispr.dbcls.jp), the potential target sites in the CT domain were retrieved and prioritized with 50%-70% GC content and an N20(or19)-NGG design. After concatenating the target site (N20(or19)) to the 5' end of an 83-bp sgRNA(Table 1), the secondary structure of the targetsgRNA sequence was predicted by RNA folding form(http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3). Only target-sgRNAs that produced less than 8-bp perfect hairpins at the target site were selected to optimize their binding to a specific site in the barleyACC1 gene. By searching for homology in the entire barley genome, the target-sgRNAs that could introduce off-target gene editing were also excluded.Using these principles, two targets that spanned about 700 bp in the CT domain were selected (Fig. 1a). The target-specific oligonucleotides are listed in Table 1.

Table 1 DNA oligonucleotides used for this research
According to an established protocol of Maet al.(2015), a single construct with Cas9 and targetsgRNA cassette was prepared. The target sequences were integrated into the single-guide RNA (sgRNA)vectors, pYLgRNA-OsU3 and pYLgRNA-OsU6a/LacZ (Maet al., 2015), and then the two targetsgRNA cassettes were cloned into the Cas9 vector pYLCRISPR/Cas9-MH (Maet al., 2015). In brief,two specific oligonucleotides (U3-T3-F and U3-T3-R for T3; U6a-T4-F and U6a-T4-R for T4; Table 1)were mixed resulting in 1 μmol · L-1solution for each oligonucleotide, treated at 95℃ for 30 s, and annealed at room temperature to form a target double helix. Targets were then incorporated into the sgRNA vectors in a restriction-ligation reaction: a 10-μL reaction contained 5 U ofBsaI, 20 U of T4 DNA ligase, 1 μL of 10×BsaI buffer, 0.3 μL of 10× T4 ligase buffer, target double helix at 0.05 mmol · L-1,20 ng pYLgRNA vector (pYLgRNA-OsU3 for T3;pYLgRNA-U6a/LacZ for T4), and deionized water.Restriction and ligation reaction was performed in a thermo-cycler at 37℃ for 5 min and then 25℃ for 5 min, which was repeated five times. For each target,two PCR reactions were performed: the first PCR with U-F and U3-T3-R (U6a-T4-R) and the second PCR with gRNA-R and U3-T3-F (U6a-T4-F) for T3 (T4),respectively (Table 1). PCR was done for 28 cycles,including 95℃ for 10 s, 60℃ for 15 s and 68℃ for 20 s. Two-round PCR products of each target were mixed and diluted at tenfold. The diluted PCR mix(ca. 2 μL) was then amplified 18 cycles at 95℃ for 10 s, 60℃ for 15 s, and 68℃ for 30 s using specific primers (B1'/B2 for target T3, B2'/BL for target T4;Table 1) that incorporated tail-specificBsaI sites on both ends of the PCR product. Purified PCR products were cloned into pYLCRISPR/Cas9-MH in a 15-μL restriction-ligation reaction that included 10 unitsBsaI, 35 U T4 DNA ligase, 1.5 μL 10×BsaI buffer,1.5 μL 10×T4 Ligase Buffer, 60-80 ng pYLCRISPR/Cas9-MH vector, 20 ng T3 PCR products and 20 ng T4 PCR products. Restriction-ligation was performed in a thermo-cycler for 15 cycles (37℃, 2 min; 10℃, 3 min;20℃, 5 min). The ligation products were transformed intoE. coliDH5α. Positive clones were confirmed by colony PCR using primers SP-ML/SP-R (Table 1).Selected clones were sequenced to confirm both targetsgRNA cassettes in the Cas9 vector (Fig. 1b).
Biolistic transformation
The biolistic transformation was adapted from an established wheat protocol (Weekset al., 1993; Lvet al., 2014;) and anAgrobacterium-mediated transfer in barley (Harwoodet al., 2009). In brief, barley caryopses were harvested 14-16 days after pollination.Immature embryos, 1-2 mm in size, were transformed with the pYLCRISPR/Cas9-ACC1-sgRNA plasmid(Fig. 1b). Transformants were then put onto selection medium containing 50 mg · L-1hygromycin. Putative transgenic plants were confirmed by positive amplification of the hygromycin phosphotransferase II(hptII) gene using primers HTP-F/HTP-R (Table 1).
Confirmation of targeted mutagenesis
Targeting induced local lesions in genomes (TILLING)(Uauyet al., 2009) was used to identifyACC1 mutations, which involved the use ofCelI and the PCR primers, HvACC1-F10 and HvACC1-R3 (Table 1).Each DNA sample contained DNA from one transgenic plant and one non-transgenic wild-type plant with 1 : 1 ratio. Identified mutations were further validated by DNA sequencing. Novel sequence variants were described by the following the nomenclature rules of the Human Genome Variation Society (den Dunnenet al., 2016).
Analysis of agronomic traits
In the T1generation, tiller numbers per plant, tiller height, spike length and the total florets per spike at the maturity stage were collected. SAS 9.4 (SAS Institute Inc., Cary, NC, USA) was used to process the collected data, which involved the use of GLM and UNIVARIATE procedures.
Herbicide test
Four APP herbicides, including clethodim, haloxyfop,quizalofop-P-ethyl and sethoxydim, were tested on the barley T1plants that were homozygous and heterozygous for the Ile1878del mutation and the T1plants with homozygous wild-type alleles. The whole plants at four-leaf stage were sprayed with haloxyfop(550-fold dilution of a solution with 10.8% active ingredient), clethodim (460-fold dilution of a solution with 30% active ingredient), quizalofop-P-ethyl(100 mg · L-1standard solution) and sethoxydim(100 mg · L-1standard solution), respectively. Herbicide responses were surveyed two weeks after treatment.

Fig. 1 Gene editing of ACC1 in barley 'Golden Promise'a, Barley ACC1 contains 33 exons in coding region. Two target sites, T3 and T4 (in turquoise), were chosen in carboxyl transferase (CT) domain.Protospacer-adjacent motif (PAM) is highlighted in yellow. b, T3 and T4 target-sgRNA cassettes in pYLCRISPR/Cas9-ACC1-sgRNA plasmid. Rice U3 and U6a promoters are used to drive T3 and T4 cassettes, respectively. c, Putative transgenic plants are confirmed by amplifying hygromycin phosphotransferase II (hptII) gene. Positive (+) and negative (-) plants have been labeled. Specific bands of the size marker (M) are indicated.d, Confirmation of 3-bp deletion in T0 plants by TILLING. Plants positive (+) and negative (-) for 3-bp deletion have been labeled. e, Sequence chromatogram of ACC1 PCR products from positive T0 plants with 3-bp deletion (top panel) or 26-bp deletion (low panel). f, Demonstration of identified editing events. In this diagram, 641 bases between two target sites are omitted.
Sequence assembly
Barley varieties 'Morex' and 'Taimalpais' were used to generate high-throughput sequence reads for genomic DNA and/or cDNA. Library preparation,high-throughput sequencing and quality control were performed by the Berry Genomics Company (Beijing,China) as described before (Niet al., 2017). Ade novoassembly of theACC1 gene was done using ABySS 2.0.2 (Simpsonet al., 2009) and SPAdes 3.12(Bankevichet al., 2012).
Results
Acetyl-CoA carboxylase genes in wheat and barley
WheatACC1 encoded the plastid form of the acetyl-CoA carboxylase (Chalupskaet al., 2008). Previous studies showed that wheat had only one copy of theACC1 gene on each homoeologous set of chromosomes (Gornickiet al., 1994; Podkowinskiet al., 1996; Gornickiet al., 1997; Fariset al.,2001; Huanget al., 2002a; Huanget al., 2002b). By searching the complete genome sequence of wheat,barley and goat grass, a single copy of the plastid form of ACCase(ACC1) was identified, which was located on the second chromosome per homologous group. The wheatACC1 had 33 exons in the coding region and encoded a 2311-aa ACCase that targeted chloroplasts (Fig. 1a). Due to its large size (>6 936 bp), the wheat sequence AF029895 in GenBank was the only full-length cDNA of theACC1 gene from Triticeae. A full-lengthACC1 DNA contig (GenBank: MK481069) was thus assembled from barley 'Tamalpais' and two full-lengthACC1 coding sequences were generated from barley 'Morex'(GenBank: MK481065) and 'Tamalpais' (GenBank:MK481066). The comparison of genome and cDNA sequences, using both the current and the published data (IBSC-v2_chr2H:19812281..19835250; (IBSC,2012), confirmed there were 33 exons in the coding region of the barleyACC1 gene, which encoded a 2311-aa ACCase. Wheat and barley ACC1 proteins displayed 98% sequence identity.
The ACC1 protein in Triticeae was characterized by six conserved domains, consisting of biotin carboxylase N-terminus, a carbamoyl-phosphate synthase L chain, a biotin carboxylase C-terminus, a biotin-requiring enzyme, an acetyl-CoA carboxylase, a carboxyl transferase (CT). The CT domain was critical for rendering the ACC1 protein sensitive or insensitive to specific herbicides (Nikolskayaet al., 1999). To date, eight residues in ACC1 had been reported to confer herbicide resistance (Table 2). By comparing to the genes of the selected Triticease species, it was found that all the eight residues were fully conserved in wheat, barley and goat grass (Table 2). For convenience, the location of critical residues inAlopecurus myosuroides(a frequently used reference for ACC1 protein) and in Triticeae was summarized (Table 2).In this study, the wheat and barley residues were recorded using their locations in the Triticeae reference.The Morex ACC1 from the International Barley Sequencing Consortium (IBSC, (IBSC, 2012)) had a W2015* truncation, but it was missing in the current cDNA assembly (MK481065) and in another Morex assembly (MLOC_52767) by the James Hutton Institute, Scotland, UK. It was possible that the Morex lines, one sequenced by IBSC and the other one from the National Small Grains Collection (# CIho 15773),were not genetically identical. However, it couldn't rule out that the current IBSC_v2 reference genome contained unintended errors in the gene region.
Editing ACC1 gene in barley 'Golden Promise'
The CT domain was critical for introducing herbicide resistance (Nikolskayaet al., 1999). Designed two CRISPR target sites, T3 and T4, which spanned about 700 bp (or 235 aa: Ala1876_Asp2110) in the CT domain and contained seven critical residues related to herbicide responses (Table 2). T3 and T4, under the control of the rice U3 and U6a promoters, respectively,were then integrated into a single CRISPR/Cas9-ACC1-gRNA plasmid (Fig. 1b). Using a PDS1000/He biolistic system, 800 immature embryos of the barley 'Golden Promise' were bombarded. Fiftyfour seedlings from eight calli survived against the hygromycin selection during regeneration, and 51 plants from eight calli were positive for the hygromycin B phosphotransferase gene (hpt) (Fig. 1c).Using TILLING, a distinctACC1 allele in eight T0plants was detected (Fig. 1d). Then, sequenced the 911-bp target region of putatively edited plants and identified two deletion patterns: a 3-bp deletion(CATC, either starting from 5' C or ending at 3' C) in the T3 site in seven plants all from one callus and a 26-bp deletion (GGTCAGAGGAACTCCAAGAGTGCAT GGG, either starting from 5' GG or ending at 3' GG)in the T4 site in one plant (Fig. 1e). All the eight T0edited plants were heterozygous for the edited bases.
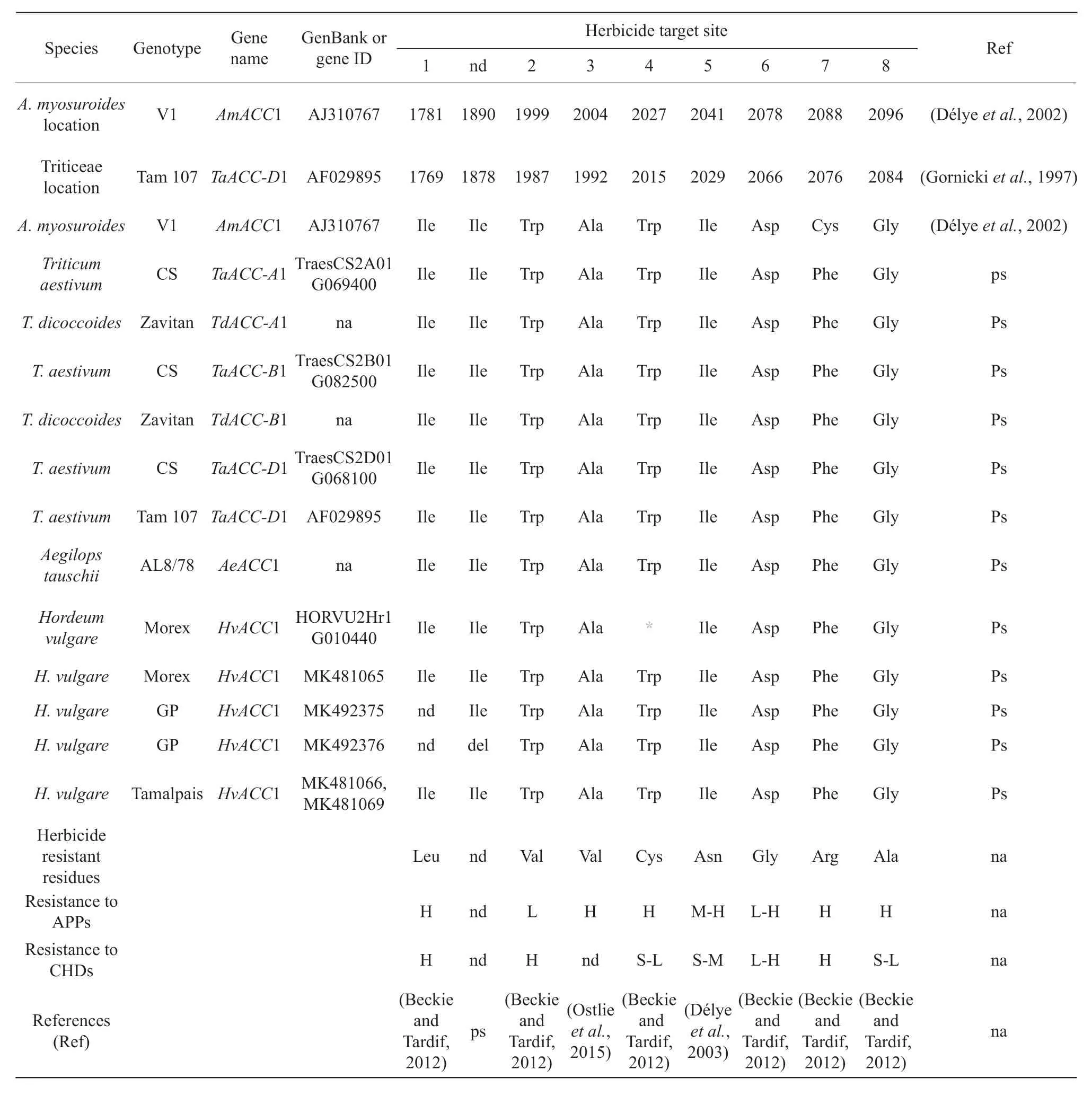
Table 2 Herbicide responsive residues in grass ACC1 protein
The 3-bp deletion at T3 resulted in the loss of Ile1878 in theACC1 gene, which in turn introduced a protein variant as ACC1:p.Ile1878del. This Ile1878 residue was conserved among all the investigated species includingA. myosuroides(Table 2). However,the 26-bp deletion at T4 caused a 213aa truncation of the C terminus, resulting in a greater variant as ACC1:p.Ser2099_Lys2311del. Considering thatACC1 was an essential gene in plants, it was concerned whether these plants would survive, even as a heterozygote,with such a severe change in itsACC1 gene. Interestingly, all the plants with the Ile1878 deletion grew well and set seeds. However, the only plant with the Ser2099_Lys2311 deletion died at the extension stage(Zadoks 32, [Zadokset al., 1974)]. Apparently, the Ile1878 deletion did not impact ACCase function, but the Ser2099_Lys2311 truncation indeed disrupted the function of ACC1 to a degree that could not be compensated by the wild-type allele.
Effect of lle1878 deletion in ACC1 on barley growth
The CT domain of the ACC1 protein where the Ile1878 deletion was located was critical for carboxyl transfer.Because this part of the protein was so important,whether the Ile1878 deletion would compromise plant growth in T1plants was investigated. Three segregation groups in the T1generation were defined,including a mutant (Mu) group homozygous for the Ile1878 deletion, a heterozygous (Het) group with only one copy of the Ile1878 deletion and a wild-type(WT) group that lacked the Ile1878 deletion. Each set of plants was assessed for four agronomic traits,including the number of fertile tillers per plant (a spike with at least five kernels), the average tiller height,the spike length and spikelets per spike. All the data met assumptions forANOVA (Levene's test,P≥0.05;Shapiro-Wilk test,P≥0.08). All the comparisons were made at the significance level of alpha=0.01.It was found that the Mu (=3.2) and Het (=5) groups were comparable to the WT (=4.8) group in fertile tillers per plant (Fig. 2a), and that the Mu (=62.2 cm)and Het (=61.9 cm) groups were similar to the WT (=62.9 cm) group in the average tiller height(Fig. 2b). Then two spike-related traits were compared: 1) the Mu (=9 cm) and Het (=8.8 cm) groups were comparable to the WT (=8.9 cm) in the spike length (Fig. 2c), and 2) the Mu (=28.8) and Het(=30) groups were similar to the WT (=29.6) group in the spikelets per spike (Fig. 2d). Apparently, the ACC1:p.Ile1878del protein was fully functional in barley, which was essential for introducing herbicide insensitivity.
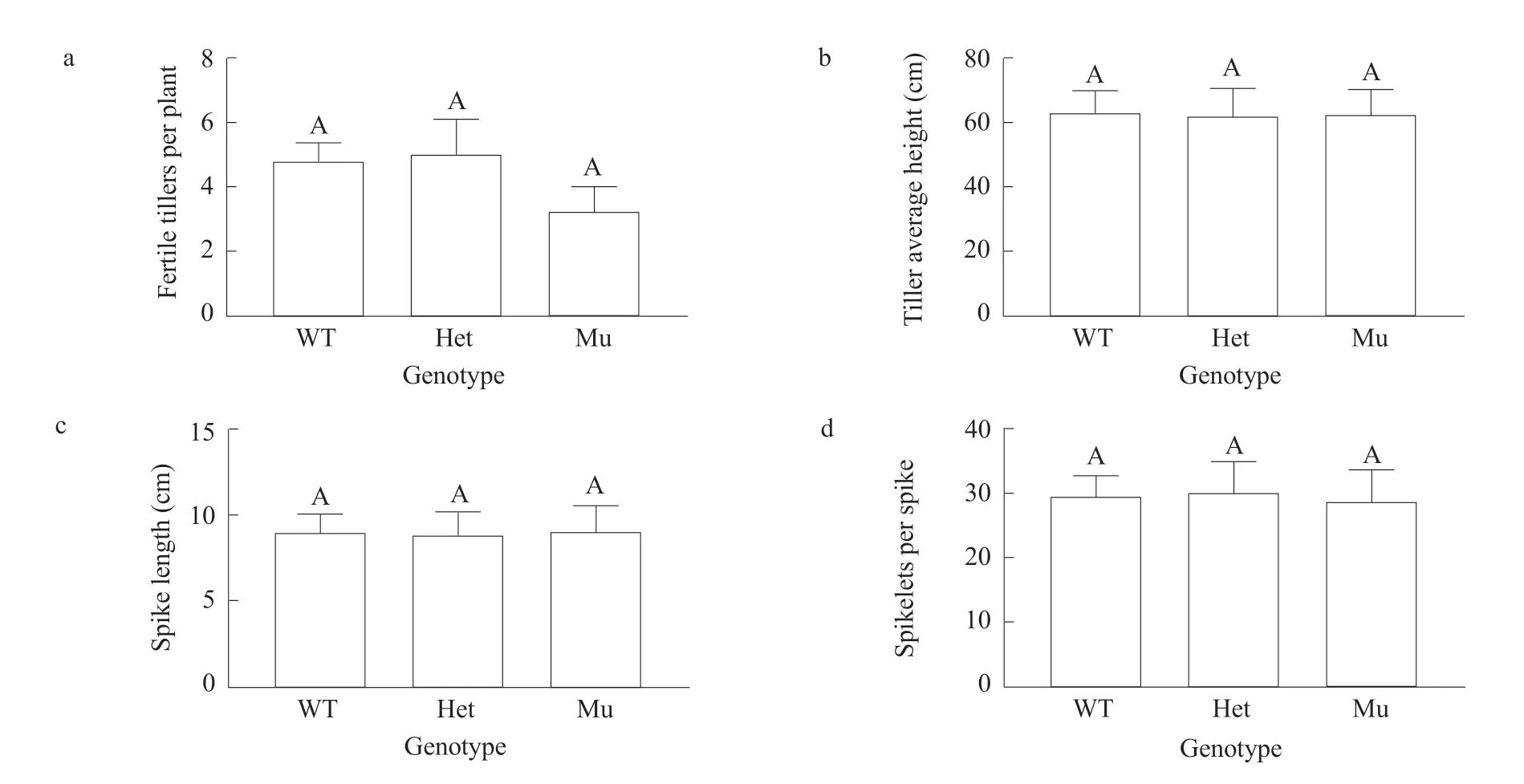
Fig. 2 Impact of Ile1878 deletion on barley growthFour agronomic traits are compared, including fertile tillers per plant (a), tiller height (b), spike length (c) and spikelets per spike (d). Three genetic groups are defined in T1 generation, including a mutant (Mu) group that is homozygous for Ile1878 deletion, a heterozygous (Het) group that is heterozygous for Ile1878 deletion, and a wild-type (WT) group that does not have Ile1878 deletion. Comparisons are made using Duncan multiple range tests at a significance level of 0.01. Error bars represent standard error of mean. Sample sizes (n) are from 6 to 9 (a) and from 19 to 43 (b-d).
Reaction of ACC1:p.lle1878del protein to four APP herbicides
To test the effect of the Ile1878 deletion, T1edited plants were treated using four selected APP herbicides:clethodim, haloxyfop, quizalofop-p-ethyl and sethoxydim. The T1plants, both homozygous and heterozygous for the Ile1878 deletion, were sensitive to all the four selected herbicides, which were comparable to the T1plants with homozygous wild-type alleles(Fig. 3).
Surprisingly, sethoxydim even caused more severe damage in plants with the Ile1878 deletion. The results showed that the ACC1:p.Ile1878del protein did not confer resistance to the selected APP herbicides.Although not tested, it was speculated that the ACC1:p.Ile1878del protein was sensitive to the CHD or other ACCase inhibitors.

Fig. 3 Reaction of ACC1-edited barley to APP herbicideSelected herbicides are tested on T1 whole plants of three genetic groups: a mutant (Mu) group that is homozygous for Ile1878 deletion, a heterozygous(Het) group that is heterozygous for Ile1878 deletion and a wild-type (WT) group that does not have Ile1878 deletion.
Discussion
ACCase catalyzed the first step in plant lipid biosynthesis, producing malonyl co-enzyme A (Malonyl-CoA) that was the only source of Malonyl-CoA in plants (Sasaki and Nagano, 2004). As a key enzyme in lipid biosynthesis, the plastid form of ACCase(or ACC1) in grass species had been reported as a major target for inducing herbicide resistance(Délye and MICHEL, 2005; Yuet al., 2007; Kukorelliet al., 2013; Tanget al., 2014). Using a yeast system,Nikolskayaet al. (1999) demonstrated that the CT domain of the wheat ACC1 protein contained the major determinant for herbicide sensitivity. To date,known herbicide resistance mutations (Beckie and Tardif, 2012; Délye, 2017) were located to the CT domain (a 316-aa fragment from Ile1769 to Gly2084 in Triticeae). It was likely that resistance to other herbicides including APP and CHD could arise from mutations in this region, but further tests would need to be done to assess which residue could be safely altered.
ACC1 was an essential and fundamental gene in lipid metabolism. Any large fragment variation might be lethal to the host plant. In the current study, the CRISPR/Cas9 construct had two separated target sites that spanned about 700 bp in the CT domain. However,no large deletions, especially those spanning the two target sites, were discovered. Even a comparatively small deletion of 26-bp appeared to be lethal to the edited plant. In this study, the gene editing rate was about 16% among all the transgenic T0plants. This was lower than reported in another study in the barley'Golden Promise' (Kapusiet al., 2017). It might be theACC1 gene itself that led to the seemingly low editing rate in the current study.ACC1 was an essential gene. Any undesirable change of this gene, especially if it occurred on both homozygous chromosomes,could be lethal and reduced the chance to recover all of the successfully edited plants that were initially generated.
Based on the limited experience gained in the present study, aiming at generating herbicide resistant grains to focus on substituting the wild-type allele with a point mutation, or to introduce an ACC1 variant with a deletion (or insertion) of one or a few residues that might be insensitive to specific herbicide groups might be best for future efforts. Up until now,a number of studies had demonstrated that it was feasible to accomplish gene substitution in plants.For example, Liet al. (2016) used two cleavage sites coupled with an engineered DNA template to substitute the riceEPSPSgenevianon-homologous end joining (NHEJ) and obtained herbicide resistant plants. In barley, NHEJ-based gene replacement with theACC1 gene was also tried. However, no edited plants were obtained (unpublished data). In contrast, using homology dependent recombination(HDR)-mediated gene replacement, Liet al. (2013)substituted the tobaccoPDSgene, Sunet al. (2016)replaced the rice acetolactate synthase (ALS), Liet al.(2018) replaced a riceNRT1.1Bgene and Wanget al.(2017) incorporated exogenous genes to a specific chromosome location in rice. Therefore, HDR-mediated gene replacement was likely to prove to be a better approach to alter the endogenousACC1 gene in barley.
It was intriguing to wonder whether an ACC1 protein with a single amino acid deletion or insertion in the CT domain would be fully functional. In this study, it was demonstrated that a single CRISPR/Cas9 plasmid with two independent target sites introduced single mutation events per plant in one of the two selected target sites. The 3-bp deletion (or Ile1878del)did not impact plant growth in the tested tiller and spike traits. Acquiring functional ACC1 variants was the first step towards introducing herbicide insensitivity in grass species. Unfortunately, the ACC1:p.Ile1878del protein was sensitive to the tested APP herbicides. A similar approach would be used to introduce other ACC1 variants, especially those close to known mutation sites that had been shown to confer herbicide resistance. Any novel mutation based on one or a few amino acids' deletions, if they could disrupt ACC1's sensitivity to APP, CHD or other ACCase inhibitors, it could be used for introducing novel herbicide resistance to specific herbicides.
Conclusions
CRISPR/Cas9 was practical to generate single amino acid deletions in the plastid localized acetyl-CoA carboxylase gene (ACC1) in barley. The Ile1878 deletion in the ACC1 protein showed no detrimental effect to the edited plants in tiller numbers, tiller height, spike length and spikelet numbers. However,the ACC1:p.Ile1878del was sensitive to the tested aryloxyphenoxypropionates (APPs) herbicides. In conclusion, this study provided a reference for barley gene editing by CRISPR/Cas9.
Author contribution statement
D.F. conceived the project; F. N. and J. W. contributed ideas; B.L., M.L., M.S. and F.J. performed the experiments; D.F and M.L. analyzed the data; D.F.,M.L. and A.C. wrote the manuscript.
Acknowledgment
We thank Dr. Yaoguang Liu of the South China Agricultural University for providing the pYLCRISPR/Cas9-MH and pYLgRNA-OsU3(U6a)/LacZ vectors.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Evaluation of Related Traits of GmTST2.1 and ZmGIF1 Genes in Soybean (Glycine Max)
- Soybean Leaf Morphology Classification Based on FPN-SSD and Knowledge Distillation
- A Bicycle Tourism Based Study on Planning and Designing of Urbanrural Recreational Greenway
—— A Case Study of Harbin City - Effects of Two Chelating Agents on Availability of Calcium and Phosphorus in Black Soil of Vegetable Fields
- Effects of Crop Rotation and Microbial Fertilizer on Nutrient Absorption and Beneficial Bacterium Abundance in Rhizosphere of Continuous Cropped Eggplant
- Effect of Chromium Propionate Substituting 25% Rumen-protected Choline on Production Performance and Blood Indicators of Perinatal Dairy Cows
