烟草(云烟87)NBS-LRR类同源抗病基因克隆及分析
2020-12-11童文杰蔺忠龙郑元仙何元胜蔡永占邓小鹏
童文杰 蔺忠龙 郑元仙 何元胜 蔡永占 邓小鹏


摘要:【目的】克隆并分析煙草NBS-LRR类抗病基因的同源序列(RGAs),为采用同源克隆技术挖掘烟草抗病基因提供理论参考。【方法】从基于NBS-LRR类抗病基因保守序列设计的简并引物中筛选扩增效果较好的引物用于扩增烟草RGAs,采用DNAMAN 6.0翻译成氨基酸序列,并与已知的其他植物抗病基因进行聚类分析。【结果】克隆获得6条与参比抗病基因具有高度同源性的烟草RGAs,大小在500 bp左右,但仅有4条RGAs具有连续的开放阅读框(ORF)及编码NBS功能结构域的核苷酸序列,命名为TRGA-87-1~TRGA-87-4。这4个烟草RGAs编码的氨基酸序列均含有NBS类抗病蛋白的多个典型保守基序,与已知的多种植物抗病基因编码的氨基酸序列均具有较高的相似性,其中,与烟草相关抗病基因编码的氨基酸序列相似性最高,为97.00%~100.00%,与其他植物的抗病基因编码氨基酸序列相似性为29.00%~53.00%,这些序列可分为3个亚类,其中TRGA-87-2与烟草N和亚麻L6聚为一类(TRGAⅠ),属于TIR-NBS-LRR类;TRGA-87-3和TRGA-87-4与水稻Xa-1和番茄Mi聚为一类(TRGAⅡ),属于non-TIR-NBS-LRR类,TRGA-87-1与拟南芥RPM1聚成一类(TRGAⅢ),属于non-TIR-NBS-LRR类。【结论】同源扩增技术可用于烟草中抗病基因的克隆、功能分析及定位等研究。
关键词: 烟草;NBS-LRR;抗病基因;同源序列;克隆
中图分类号: S572 文献标志码: A 文章编号:2095-1191(2020)09-2138-07
Cloning and analysis of NBS-LRR resistance gene
analogs in tobacco(Yunyan 87)
TONG Wen-jie1, LIN Zhong-long2, ZHENG Yuan-xian3, HE Yuan-sheng3,
CAI Yong-zhan4, DENG Xiao-peng1*
(1Yunnan Academy of Tobacco Science, Kunming 650201, China; 2Yunnan Branch of China National Tobacco Corporation, Kunming 650011; 3Lincang Branch of Yunnan Tobacco Company, Lincang, Yunnan 677000,
China; 4Qujing Branch of Yunnan Tobacco Company, Qujing, Yunnan 655000, China)
Abstract:【Objective】To clone and analyze the resistance gene analogs(RGAs) of NBS-LRR resistance genes in tobacco,provide theoretical reference for using homologous cloning technology to mine tobacco resistance genes. 【Method】The degenerate primers designed based on the conserved sequences of NBS-LRR resistance genes were selected to amplify RGAs of tobacco. The vector sequences in the sequenced fragments were removed by DNAMAN 6.0,and then transla-ted into amino acid sequences. Homology analysis was performed in NCBI database,and cluster analysis was performed with other known disease resistance genes. 【Result】Six tobacco RGAs with high homology with the reference disease resistance genes were cloned and their sizes were about 500 bp. Only four RGAs had continuous open reading frames (ORFs) and nucleotide sequences encoding NBS functional domain,named from TRGA-87-1 to TRGA-87-4. The amino acid sequences encoded by these four RGAs all contained several typical conserved motifs of NBS like resistance proteins,and shared high similarity with known amino acid sequences encoded by many plant disease resistance genes.Among them,the amino acid sequences of tobacco related disease resistance genes had the highest similarity(97.00%-100.00%) and 29.00%-53.00% with other plant disease resistance genes. These sequences could be divided into three subgroups. TRGA-87-2,tobacco N and flax L6 were clustered into one group(TRGA I), belonging to TIR-NBS-LRR resistance genes. TRGA -87-3, TRGA -87-4, rice Xa-1 and tomato Mi clustered into a subgroup(TRGAⅡ), belonging to non-TIR-NBS-LRR resistance genes. TRGA-87-1 and arabidopsis RPM1 clustered into a subgroup(TRGA III),belonging to non-TIR-NBS-LRR resistance genes. 【Conclusion】Homologous amplification can be used for cloning,functional analysis and localization of disease resistance genes in tobacco.
Key words: tobacco; NBS-LRR; resistance gene; homologous sequence; cloning
Foundation item: National Natural Science Foundation of China(31660426); Project of Yunnan Branch of China National Tobacco Corporation(2018530000241016,2018530000241020,2019530000241011)
0 引言
【研究意义】携带核苷酸结合位点(NBS)和亮氨酸富集重复(LRR)结构的NBS-LRR类抗病基因是植物基因组中所含抗病基因数目最多的基因家族。近年来许多植物基因组测序工作已相继完成,为从全基因组水平上研究和分析NBS-LRR类抗病基因打下基础(Meyers et al.,1999,2003;The Tomato Genome Consortium,2012;李任建等,2020)。烟草(Nicotiana tabacum L.)作为重要的经济作物之一,其基因组测序也已完成。目前病虫害的发生严重制约烟叶田间生产。虽然烟草品种自身在一定程度上对病虫害具有抵抗力(张艳云,2011),但不同烟草品种对不同病毒类型和虫害的抗性差异很大(张雪峰,2014;王家川等,2016)。因此,挖掘烟草中更多NBS-LRR类抗病基因,对各种烟草病害的防治极其重要,对烟草安全高效生产具有重要意义。【前人研究进展】目前,采用图位克隆、转座子标签和同源克隆等技术已从拟南芥、小麦、甘薯、马铃薯和甘蔗等植物中获得多种抗病基因,这些基因可激活植株抗病机制(Shirano et al.,2002;Sekhwal et al.,2015;Zhou et al.,2016;毕楚韵等,2020)。据前人研究报道,NBS-LRR类抗病基因广泛存于真核生物中,其编码蛋白主要包括Tir/CC、NBS和LRR三大结构域(图1)(刘云飞等,2014;李任建等,2020),其中,NBS保守結构域又可分为8个保守基序(Zhou et al.,2004;刘云飞等,2014)。近年来,利用NBS-LRR类抗病基因的保守序列对不同植物抗病基因进行广泛挖掘,已从不同植物中克隆得到较多该类抗病基因的同源序列(RGAs)。张立荣等(2011)根据NBS-LRR类抗病基因保守序列设计简并引物,从小麦中扩增得到13条具有开放阅读框(ORF)的RGAs;陈玲等(2012)从悬钩子蔷薇中克隆获得4条RGAs。此外,NBS-LRR类抗病基因也可用于分子标记及基因定位的开发等。Liu等(2013)从花生RGAs中挖掘出28个SSR分子标记,并将其中的RGA121标记定位连锁群AhIV上;丁玉梅(2019)根据NBS-LRR类抗病基因保守序列设计简并引物,从黑籽南瓜中克隆获得8条RGAs,并将其作为参考序列与转录组和蛋白组相结合筛选抗病基因;Kim等(2004)、Qi等(2014)研究拟南芥NBS-LRR类抗病蛋白RPS5识别方式,对多种植物的病原菌具有抗病作用。【本研究切入点】尽管在烟草上有很多关于抗病基因的研究(张立荣等,2011;袁清华等,2014;曾建敏等,2016),但其抗病基因功能及分类尚不清楚。有关烟草NBS-LRR类抗病基因的研究鲜见报道。【拟解决的关键问题】根据已报道的NBS-LRR类抗病基因保守序列设计并合成简并引物,从烟草中扩增获得RGAs,并对其序列进行分析,为利用同源克隆技术挖掘烟草抗病基因提供理论参考。
1 材料与方法
1. 1 试验材料
供试烟草品种为云烟87,具有抗黑胫病、南方根结线虫病、普通花叶病及叶斑病等抗性。烟草种子由云南省烟草农业科学研究院农艺中心提供。主要试剂:pMD18-T Vector试剂盒和大肠杆菌DH5α感受态细胞均购自宝生物工程(大连)有限公司;胶回收试剂盒购自北京全式金生物科技有限公司。主要仪器设备:PCR仪(Eppendorf Mastercycler Nexus,德国)、凝胶成像系统(VILBER Quantum CX5,法国)和紫外可见分光光度计(Implen,德国)。
1. 2 试验方法
1. 2. 1 烟草DNA提取 采集田间种植的烟草叶片,参考Allen等(2006)的CTAB法提取其总DNA,方法略有改动。
1. 2. 2 简并引物设计及筛选 前人已根据拟南芥、亚麻、甘蔗、烟草和甘薯等NBS-LRR类抗病蛋白NBS保守结构域的编码核苷酸序列设计简并引物,从中筛选出扩增效果较好的引物用于烟草RGAs克隆,其序列信息如表1所示。
1. 2. 3 烟草RGAs克隆及测序 利用提取的DNA和筛选获得的简并引物进行PCR扩增。反应体系25.0 μL:10×PCR Buffer(plus Mg2+)2.5 μL,2.5 mmol/L dNTPs 0.5 μL,10 μmol/L上游引物(1F、2F和3F)2.5 μL,10 μmol/L下游引物(1R、2R和3R)2.5 μL,Taq DNA聚合酶(5 U/μL)0.2 μL,1 μL DNA模板,ddH2O补足至25.0 μL。扩增程序、回收纯化及测序参照魏环宇等(2019)的方法。
1. 2. 4 序列分析及系统发育进化树构建 使用DNAMAN 6.0将测序获得的核苷酸序列翻译成氨基酸序列,将其提交至NCBI数据库进行BLASTx比对。用DNAMAN 6.0对已知功能的抗病基因进行多序列比对,并构建系统发育进化树。
2 结果与分析
2. 1 烟草RGAs克隆及测序结果
由图2可知,克隆获得6条与参比抗病基因具有高度同源性的烟草RGAs,长度约500 bp,与预期结果相符。序列分析结果显示,6条烟草RGAs中,有2个RGAs为不可表达的假基因,其余4个RGAs含有连续且完整的ORF及编码NBS功能结构域的核苷酸序列(GenBank登录号分别为MK634312、MK634313、MK634314和MK634315),命名为TRGA-87-1~TRGA- 87-4。可见,采用同源扩增技术可有效挖掘烟草RGAs。
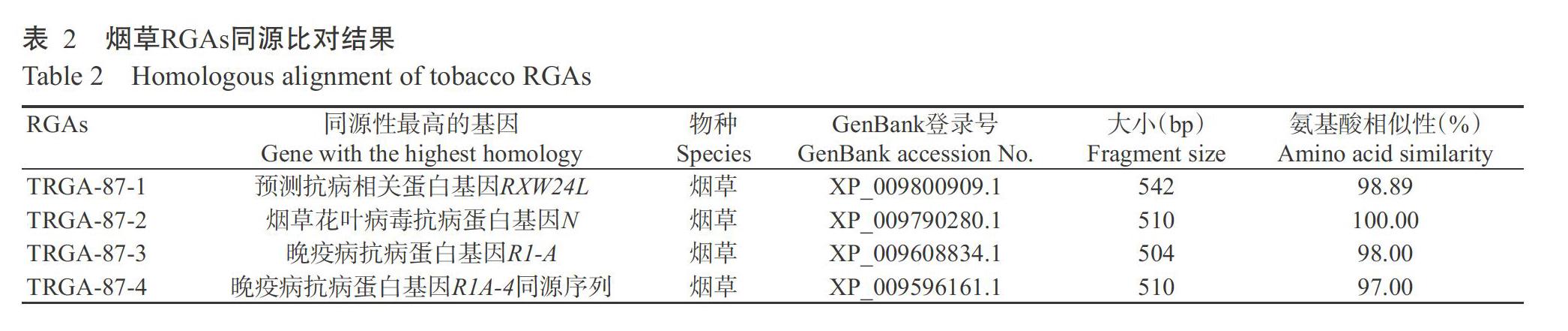
2. 2 烟草RGAs同源性分析结果
通过同源性比对分析发现,4条RGAs编码的氨基酸序列与已知的多种植物抗病基因编码的氨基酸序列均具有较高的相似性,其中,与烟草相关抗病基因编码的氨基酸序列相似性较高,为97.00%~100.00%(表2)。
2. 3 烟草RGAs编码的保守结构域分析结果
利用DNAMAN 6.0对4条烟草RGAs编码的氨基酸序列与4条其他植物抗病基因编码的氨基酸序列进行多重比对分析,结果(图3)显示,4条烟草RGAs编码的氨基酸序列均含有NBS类抗病蛋白的多个典型保守基序,即Kinase-2(LIVLDDVW)模体、P-loop(GMGGVGGKTT)和GLPLAL疏水性区段,在Kinase-2区的最后一个氨基酸为天冬氨酸或色氨酸,推测这8条RGAs可分为non-TIR-NBS-LRR(CC-NBS-LRR)类和TIR-NBS-LRR类。这4条烟草RGAs与5条已报道的抗病相关植物的RGAs编码的氨基酸序列在P-loop(GMGGVGG KTT)、Kinase-2(LIVLDDVW)和GLPLAL保守结构域中具有高度相似性。
2. 4 烟草RGAs聚类分析结果
应用DNAMAN 6.0将4条烟草RGAs编码的氨基酸序列与拟南芥、烟草、亚麻、番茄和水稻等植物抗病基因编码的氨基酸序列进行相似性分析,并构建系统发育进化树,结果(图4)显示,其氨基酸序列相似性为29.00%~53.00%,可分为3个亚类,其中,TRGA-87-2与烟草N(AAA50763)和亚麻L6(AAA91022)聚为一类(TRGAⅠ),尤其与烟草N的相似性最高,为53.00%,与亚麻L6相似性为39.00%,结合上述同源性分析结果,推测TRGA-87-2是与烟草花叶病毒有关的抗病基因,属于TIR-NBS-LRR类;TRGA-87-3和TRGA-87-4与水稻Xa-1(BAA25068)和番茄Mi(AAC9x7933)聚为一类(TRGAⅡ),其中,TRGA-87-3与TRGA-87-4相似性为67.00%,二者与番茄Mi相似性均为49.00%,与水稻Xa-1相似性均为32.00%,推测二者具有相似的基因功能,結合上述同源性分析结果,二者均为与烟草晚疫病有关的抗病基因,属于non-TIR-NBS-LRR类;而TRGA-87-1与拟南芥RPM1(X87851)聚成一类(TRGAⅢ),二者相似性为29.00%,也属于non-TIR-NBS-LRR类。可见,烟草RGAs属于NBS类抗病基因,分为non-TIR-NBS-LRR和TIR-NBS-LRR两大类,与上述保守结构域分析结果一致。
3 讨论
同源克隆技术具有操作简单、扩增模板用量少、统计效率高等优点,可应用于基因组复杂的植物中,是一种便捷、经济的分离和克隆未知基因的有效方法。本研究运用该技术获得6条烟草RGAs,其中有2条可能存在内含子,无法通读,为不表达的假基因。因此,在后续研究中可以cDNA为扩增模板,其原因是cDNA中均为表达序列,不含任何内含子,能获得准确度更高的RGAs,更有利于筛选出抗病基因(路妍等,2020)。NBS-LRR类抗病基因约占所有抗病基因的72%,通常以基因家族的形式广泛存在于植物基因组中(阙友雄等,2009;薛莹莹等,2014)。当植物受到病原菌侵染时,通过超表达NBS类抗病基因从而启动侵染部位的程序性死亡,以提高植物抗病害能力(丁玉梅,2019)。本研究利用简并引物PCR扩增获得4条烟草RGAs,通过BLAST比对发现,其编码蛋白与已报道的抗病基因编码蛋白具有较高的氨基酸序列相似性,含有蛋白激酶中的保守基序,有可能参与植物抗病反应过程中的信号识别和传导等途径,因此,这4条烟草RGAs可作为抗病相关的候选基因,进一步分析其与已知抗病基因的关系,从而挖掘出新的抗病基因。
目前,国内外在NBS-LRR类抗病基因研究方面取得较大进展,已从黄瓜(丁国华等,2005;王昶童等,2014)、芒果(刘洋等,2013)、甘薯(王连军等,2013;黄小芳等,2020)、葡萄(张颖等,2013)、核桃(安海山和杨克强,2014)、桑树(刘潮等,2019)等基因组未知的作物中,克隆得到大量NBS-LRR类RGAs,为抗病基因的研究打下基础。但NBS-LRR类抗病基因的相关研究仍不够深入。目前已从水稻、拟南芥等基因组测序完成的植物中克隆获得具有许多共同特点的NBS-LRR类抗病基因,但其抗病基因数目结构和类型存在明显差异,为了有效利用抗病基因,还需对更多植物的RGAs进行深入分析,解析其特点。由于NBS-LRR类基因家族成员众多,同时不同烟草品种对不同病虫害的抗性不同,有关烟草抗病基因的分类,乃至NBS-LRR类烟草抗病基因至今尚未系统研究分析。
4 结论
利用简并引物进行同源扩增是挖掘烟草RGAs的有效方法,可用于烟草中抗病基因的克隆、功能分析及定位等研究。
参考文献:
安海山,杨克强. 2014. 核桃NBS类抗病基因类似物的序列特征及其与炭疽病的抗性[J]. 中国农业科学,47(2):344-356. [An H S,Yang K Q. 2014. Sequence analysis of NBS-type RGAs and their relationship with anthracnose resistance in walnut[J]. Scientia Agriculture Scinica,47(2):344-356.]
毕楚韵,黄小芳,周丽香,石媛媛,胡韵卓,梁才晓,黄碧芳,许明,林世强,陈选阳. 2020. 三浅裂野牵牛NBS-LRR类抗病基因的鉴定和分析[J/OL]. 分子植物育种,https://kns.cnki.net/kcms/detail/46.1068.S.20200612.1815.012.html. [Bi C Y,Huang X F,Zhou L X,Shi Y Y,Hu Y Z,Liang C X,Huang B F,Xu M,Lin S Q,Chen X Y. 2020. Identification and analysis of NBS-LRR gene family in Ipomoea trifida genome[J]. Molecular Plant Breeding,https://kns.cnki.net/kcms/detail/46.1068.S.20200612.1815.012. html.]
陳玲,张颢,邱显钦,晏慧君,王其刚,蹇洪英,唐开学. 2012. 云南悬钩子蔷薇NBS-LRR类抗病基因同源克隆与分析[J]. 植物分类与资源学报,34(1):56-62. [Chen L,Zhang H,Qiu X Q,Yan H J,Wang Q G,Jian H Y,Tang K X. 2012. Cloning and analysis of NBS-LRR type di-sease resistance gene analogs from Rosa rubus in Yunnan[J]. Plant Diversity and Resources,34(1):56-62.]
丁国华,秦智伟,刘宏宇,周秀艳,迟春玉,王志坤. 2005. 黄瓜NBS类型抗病基因同源序列的克隆与分析[J]. 园艺学报,32(4):638-642. [Ding G H,Qin Z W,Liu H Y,Zhou X Y,Chi C Y,Wang Z K. 2005. Analysis and clo-ning of NBS class disease resistant gene analog in cucumber[J]. Acta Horticulturae Sinica,32(4):638-642.]
丁玉梅. 2019. 黑籽南瓜对枯萎病菌侵染的应答机制及NBS类抗病基因筛选[D]. 重庆:西南大学. [Ding Y M. 2019. The response mechanism of Cucurbita ficifolia infected by Fusarium oxysporum f. sp. Cucumerinum and selecting of NBS type disease-resistance genes[D]. Chong-qing:Southwest University.]
黄小芳,毕楚韵,石媛媛,胡韵卓,周丽香,梁才晓,黄碧芳,许明,林世强,陈选阳. 2020. 甘薯基因组NBS-LRR类抗病家族基因挖掘与分析[J]. 作物学报,46(8):1195-1207. [Huang X F,Bi C Y,Shi Y Y,Hu Y Z,Zhou L X,Liang C X,Huang B F,Xu M,Lin S Q,Chen X Y. 2020. Discovery and analysis of NBS-LRR gene family in sweet potato genome[J]. Acta Agronomica Sinica,46(8):1195-1207.]
李任建,申哲源,李旭凯,韩渊怀,张宝俊. 2020. 谷子NBS-LRR类基因家族全基因组鉴定及表达分析[J]. 河南农业科学,49(2):34-43. [Li R J,Shen Z Y,Li X K,Han Y H,Zhang B J. 2020. Genome-wide identification and expression analysis of NBS-LRR gene family in Setaria italica[J]. Journal of Henan Agricultural Sciences,49(2):34-43.]
刘潮,褚洪龙,韩利红,杨云锦,高永,唐利洲. 2019. 桑树NBS-LRR类基因家族的全基因组鉴定及其调控microRNAs分析[J]. 江苏农业学报,35(3):544-553. [Liu C,Chu H L,Han L H,Yang Y J,Gao Y,Tang L Z. 2019. Genome-wide identification of NBS-LRR genes and regulation analysis by microRNAs in mulberry[J]. Jiangsu Journal of Agricultural Sciences,35(3):544-553.]
刘洋,姚全胜,苏俊波,洪亚楠,雷新涛. 2013. 芒果NBS类抗病基因同源序列克隆与分析[J]. 植物遗传资源学报,14(3):571-576. [Liu Y,Yao Q S,Su J B,Hong Y N,Lei X T. 2013. Isolation and characterization of NBS type resistance gene analogs from mango(Mangifera indica Linn.)[J]. Journal of Plant Genetic Resources,14(3):571-576.]
刘云飞,万红建,李志邈,叶青静,王荣青,阮美颖,姚祝平,周国治,韦艳萍,杨悦俭. 2014. 植物NBS-LRR抗病基因的结构、功能、进化起源及其应用[J]. 分子植物育种,12(2):377-389. [Liu Y F,Wan H J,Li Z M,Ye Q J,Wang R Q,Ruan M Y,Yao Z P,Zhou G Z,Wei Y P,Yang Y J. 2014. Analysis of plant NBS-LRR resistance gene:Structure,function,origin,evolution and their application[J]. Molecular Plant Breeding,12(2):377-389.]
路妍,刘洋,宋阳,景岚. 2020. 向日葵NBS-LRR抗病基因家族全基因组分析[J]. 中国油料作物学报,42(3):441-452. [Lu Y,Liu Y,Song Y,Jing L. 2020. Genome-wide analysis of NBS-LRR-encoding gene in Helianthus an-nuus[J]. Chinese Journal of Oil Crop Sciences,42(3):441-452.]
阙友雄,许莉萍,林剑伟,陈如凯. 2009. 甘蔗NBS-LRR类抗病基因同源序列的分离与鉴定[J]. 作物学报,35(4):631-639. [Que Y X,Xu L P,Lin J W,Chen R K. 2009. Isolation and characterization of NBS-LRR resistance gene analogs from sugarcane[J]. Acta Agronomica Sinica,35(4):631-639.]
王昶童,曹刚强,梁从敏,谢冰心,闫飞翔. 2014. 黄瓜NBS类抗病基因类似物的生物信息学分析[J]. 北方园艺,(14):101-104. [Wang C T,Cao G Q,Liang C M,Xie B X,Yan F X. 2014. Bioinformatics analysis of the NBS resistance gene analogs in cucumber[J]. Northern Horticulture,(14):101-104.]
王家川,吳国贺,冯月,李久道,朴世领. 2016. 33份烟草品种(系)TMV病鉴定与评价分析[J]. 吉林农业大学学报,38(6):656-662. [Wang J C,Wu G H,Feng Y,Li J D,Piao S L. 2016. Identification and evaluation of resistance of 33 tobacco varieties(lines) to TMV disease[J]. Journal of Jilin Agricultural University,38(6):656-662.]
王连军,贾礼聪,苏文瑾,雷剑,杨新笋. 2013. 甘薯近缘野生种抗病基因同源序列的分离和鉴定[J]. 湖北农业科学,52(11):2680-2683. [Wang L J,Jia L C,Su W J,Lei J,Yang X S. 2013. Isolation and characterization of resistance gene analogs from wild relative of sweet potato[J]. Hubei Agricutural Sceinces,52(11):2680-2683.]
魏环宇,童文杰,蔺忠龙,莫笑晗,张丽芳,何元胜,郑元仙,王继明,许银莲,陈小龙,钟宇,余磊,邓小鹏. 2019. 烟草(红花大金元)NBS-LRR类抗病基因同源序列的克隆与分析[J]. 西南农业学报,32(12):2747-2751. [Wei H Y,Tong W J,Lin Z L,Mo X H,Zhang L F,He Y S,Zheng Y X,Wang J M,Xu Y L,Chen X L,Zhong Y,Yu L,Deng X P. 2019. Isolation and analysis of NBS-LRR resistance gene analogs from tobacco(Hongda)[J]. Southwest China Journal of Agricultural Sciences,32(12):2747-2751.]
薛莹莹,孙守如,孙德玺,邓云,朱迎春,刘君璞. 2014. RGA法克隆NBS-LRR类抗病基因同源序列及其在葫芦科作物上应用的研究进展[J]. 中国瓜菜,27(3):1-4. [Xue Y Y,Sun S R,Sun D X,Deng Y,Zhu Y C,Liu J P. 2014. Cloning of NBS-LRR-encoding resistance gene analogues using RGA approach and the advance in cucurbit crops[J]. China Cucurbits and Vegetable,27(3):1-4.]
袁清华,谢锐鸿,张振臣,马柱文,李集勤,李淑玲,陈俊标. 2014. 烟草表达抗病基因同源物(RGA)的鉴定及RGA-SSR标记的开发[J]. 作物学报,40(2):240-252. [Yuan Q H,Xie R H,Zhang Z C,Ma Z W,Li J Q,Li S L,Chen J B. 2014. Identification of expressed resistance gene analogues(RGAs) and development of RGA-SSR markers in nicotiana[J]. Acta Agronomica Sinica,40(2):240-252.]
曾建敏,吴兴富,李梅云,张谊寒,焦芳婵,李永平. 2016. 烤烟品种NC196抗性分子检测及特征特性分析[J]. 分子植物育种,14(10):2829-2836. [Zeng J M,Wu X F,Li M Y,Zhang Y H,Jiao F C,Li Y P. 2016. Molecular identification of disease resistance and characteristics of a fluecured tobacco variety NC196[J]. Molecular Plant Bree-ding,14(10):2829-2836.]
张立荣,杨文香,刘大群. 2011. 小麦NBS类抗病基因类似序列的多样性和进化关系研究[J]. 华北农学报,26(4):23-26. [Zhang L R,Yang W X,Liu D Q. 2011. Diversity and evolutionary relationship of NBS-type resistance gene analogues in wheat[J]. Acta Agriculturae Boreali-Sinica,26(4):23-26.]
张雪峰. 2014. 烟草抗TMV突变体抗性遗传分析与相关基因鉴定[D]. 北京:中国农业科学院. [Zhang X F. 2014. Genetic analysis of resistance and identification of relative genes in TMV-resistant tobacco mutants[D]. Beijing:Chinese Academy of Agricultural Sciences Dissertation.]
张艳云. 2011. 抗花叶病烟草种质资源的筛选[D]. 福州:福建农林大学. [Zhang Y Y. 2006. Screening of tobacco germplasm resourses resisted Tobacoo Mosaic Virus[D]. Fuzhou:Fujian Ageiculture and Forestry University.]
張颖,李峰,刘崇怀,樊秀彩,孙海生,姜建福,张国海. 2013. 中国野生刺葡萄抗白腐病NBS-LRR类抗病基因同源序列的分离与鉴定[J]. 中国农业科学,46(4):780-789. [Zhang Y,Li F,Liu C H,Fan X C,Sun H S,Jiang J F,Zhang G H. 2013. Isolation and identification of NBS-LRR resistance gene analogs sequences from Vitis davidii[J]. Scientia Agricultura Sinica,46(4):780-789.]
Allen G C,Flores-Vergara M A,Krasynanski S. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide[J]. Nature Protocols,1(5):2320-2325.
Deng Z,Huang S,Ling P,Chen C,Yu C,Weber C A,Moore G A,Gmitter G A. 2000. Cloning and characterization of NBS-LRR class resistance-gene candidate sequences in citrus[J]. Theoretical and Applied Genetics,101(5):814-822.
Kanazin V,Laura F M,Randy C S. 1996. Resistance gene ana-logs are conserved and clustered in soybean[J]. Trends in Genetics,13(2):54.
Kim S T,Kim S G,Hwang D H. 2004. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus[J]. Proteomics,4(11):3569-3578.
Leister D,Ballvora A,Francesco S,Gebhardt C. 1996. A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants [J]. Nature Publishing Group,14(4):421-429.
Liu Z J,Feng S,Pandey M K,Chen X,Culbreath A K,Varshney R K,Guo B Z. 2013. Identification of expidentification of expressed resistance gene analogs from peanut(Arachis hypogaea L.) expressed sequence tagss[J]. Journal Integrative Plant Biology,55(5):453-461.
Meyers B C,Dickerman A W,Michemore R W,Sivaramakrishnan S,Sobral B W,Young N D. 1999. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily[J]. The Plant Journal,20(3):317-332.
Meyers B C,Kozik A,Griego A,Kuang H H,Michelmore R W. 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis[J]. Plant Cell,15(4):809-834.
Qi D,Dubiella U,Kim S H. 2014. Recognition of the protein kinase avrpphb susceptible 1 by the disease resistance protein resistance to Pseudomonas syringae 5 is dependent on s-acylation and an exposed loop in avrpphb susceptible 1[J]. Plant Physiology,164(1):340-351.
Sekhwal M K,Li P C,Lam I,Wang X,Cloutier S,You F M. 2015. Disease resistance gene analogs(RGAs) in plants[J]. International Journal of Molecular Sciences,16(8):19248-19290.
Shirano Y,Kachroo P,Shah J,Klessig D F. 2002. Gain-of-function mutation in an arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance[J]. Plant Cell,14(12):3149-3162.
The Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution[J]. Nature,485(7400):635-641.
Zhou F,Guo Y,Qiu L J. 2016. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean[J]. BMC Plant Bio-logy,16:58-71.
Zhou T,Wang Y,Chen J Q,Araki H,Jing Z,Jiang K,Shen J,Tian D. 2004. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes[J]. Molecular Genetics and Genomics,271(4):402-415.
(責任编辑 陈 燕)
