Taxonomy and Phylogeny of a New Marine Planktonic Ciliate, Strombidium pseudorapulum sp. n.(Protozoa, Ciliophora, Oligotrichia)
2020-09-28LIFenfenHUANGYaoyaoYUYingLIUWeiweiandLINXiaofeng
LI Fenfen HUANG Yaoyao YU Ying LIU Weiwei and LIN Xiaofeng 5)
Taxonomy and Phylogeny of a New Marine Planktonic Ciliate,sp. n.(Protozoa, Ciliophora, Oligotrichia)
LI Fenfen1), HUANG Yaoyao1), YU Ying1), LIU Weiwei2), 3), 4), *, and LIN Xiaofeng1), 5),*
1),,,,510631,2),,,, 510301,3),, 511458,4),,510301,5),,,361005,
As one of the main groups of marine planktonic protozoa, oligotrich ciliates have shown a high biodiversity. Recently, a new aloricate oligotrich ciliate,sp. n. was isolated from coastal waters of southeast China. The living morphology and infraciliature of this species were studied using live cell observation and protargol impregnation.sp. n. differs from its congeners by having a radish-like body with a bumped anterior end and an elongated tail-like posterior end, possessing a red ‘eyespot’ in apical protrusion of cell, adoral zone of membranelles composed of 12–16collar and 9–12 buccal membranelles, one spindle-shaped macronucleus, girdle kinety in anterior 2/5 of cell with 45–60 densely spaced dikinetids, and ventral kinety occupying about the posterior 1/3 of the cell with 16–19 dikinetids.sp. n. clusters together with those species having ‘eyespot’ in apical protrusion in the phylogenetic tree based on the small subunit rRNA gene sequence. It is a sister species ofwith 31 unmatched nucleotides between their SSU rRNA gene sequences.
aloricate oligotrichs; morphology; new species; SSU rRNA gene sequence
1 Introduction
As one of the dominant groups of microzooplankton communities, aloricate oligotrich ciliates have been stud- ied worldwide for a century and a half in terms of tax- onomy, and about 200 species have been reported (Song, 2009; Agatha, 2011; Liu, 2017, 2019). Recent studies have demonstrated that this assemblage is much more diverse than previously supposed (Liu, 2011, 2013, 2016; Hu, 2019; Song, 2018).
species are very common oligotrich cili- ates in marine and fresh waters. Characterized by hori- zontal girdle kinety and longitudinal ventral kinety, the genuswas established by Claparède and Lach- mann (1859). During the past 14 years, 17 newspecies have been reported, which indicates that there is still much to learn about the diversity of this genus (Agatha, 2005; Song, 2005; Wilbert and Song, 2005; Xu, 2008; Song, 2009; McManus, 2010; Song, 2015a, b; Liu, 2015, 2016; Elangovan and Gauns, 2017).Furthermore, the phylogenetic rela- tionships ofspecies were still not well ad- dressed since the monophyly of the genus was not sup- ported in previous studies (Gao, 2009; Kim, 2010; Li, 2013; Agatha and Strüder-Kypke, 2014; Song, 2018). With the continuous supplement of mole- cular markers, the phylogenetic relationships of this group could be further investigated (Gao, 2016). There is a clear need for further taxonomic studies on this genus using a combination of morphological and molecular ap- proaches.
During a study on the biodiversity of marine plankto- nic ciliates in coastal waters of China, a new aloricate oli- gotrich species,sp. n. was found near a wharf of Fujian Province. Its morphological characters were investigated based on live observations and silver staining. In addition, its phylogenetic position was analyzed based on SSU rRNA gene sequences.
2 Materials and Methods
2.1 Sample Collection and Morphological Study
sp. n. was sampled from coastal waters near the Songyu Wharf, Xiamen (118˚05΄E, 24˚45΄N), Fujian Province on May 19, 2018. Water sam- ples were collected using 100mL bottles at sampling site. The samples were then transferred into the Petri dishes and the specimens were immediately isolated for further investigation in the lab. The morphology of living cells were studied using bright-field and differential interfere- ence contrast microscopy (Nikon 80i). The infraciliature was revealed through protargol impregnation (Wilbert, 1975). Drawings of live cells were based on photomicro- graphs, while those of silver-impregnated cells were made using a camera lucida (×1000). Counts and measurements were performed at a magnification of ×1000. Terminol- ogy accords to Agatha and Riedel-Lorjé (2006). Classi- fication follows Lynn (2008) and Adl(2019).
2.2 Extraction, Amplifcation and Sequencing of DNA
A single individual from the raw culture was harvested, washed several times with filteredwater, and placed in a 1.5mL centrifuge tube. Genomic DNA was extracted using a DNeasy Blood & Tissue kit (Qiagen, CA) following the manufacturer’s instructions. The PCR primers used to amplify the SSU rRNA gene followed the method of Medlin(1988), and Q5 Hot Start High-Fidelity DNA Polymerase (New England BioLabs, USA) was used in the experiment. Cycling parameters were as follows: 30s initial denaturation (98℃), 35 cy- cles of 10s at 98℃, 30s at 56℃, and 30s at 72℃ with a final extension of 10min at 72℃. PCR products were se- quenced in Guangzhou Tianyi Huiyuan Gene Inc.
2.3 Phylogenetic Analyses
In addition tothat ofsp. n. from this study, the SSU rRNA gene sequences of 70 ciliate species retrieved from GenBank database include- ing all genus of the family Strombidiidae were used to construct the phylogenetic trees; two species of Hypotri- chia were used as the outgroup taxa. All sequences were aligned using Bioedit (Hall, 1999) with default parame- ters, and the ends of alignments were trimmed yielding a matrix of 1757 characters. Maximum likelihood (ML) ana- lyses were conducted using RaxML-HPC2 on XSEDE (8.2.12) (Stamatakis, 2006; Stamatakis, 2008) with the GTRGAMMA model provided on the online server CIPRES Science Gateway (Miller, 2010). The reli- ability of internal branches was assessed using a non- parametric bootstrap method with 103replicates, and search- es for the best tree were conducted starting from 1000 random trees, then multiparametric bootstrapping was con- ducted to get accurate support value from these 1000 trees consensus. Bayesian inference (BI) analysis was perform- ed using MrBayes 3.1.2 on XSEDE v 3.1.2 (Ronquist and Huelsenbeck, 2003) provided on the CIPRES Sci- ence Gateway with the model GTR+I+G selected by the Akaike Information Criterion (AIC) in MrModeltest v2 (Nylander, 2004). Markov chain Monte Carlo analyses were run for 6×106generations with two parallel runs, each with four simultaneous chains, sampling every 100 generations. The first 15000 trees were discarded as burn- in. The remaining trees were used to calculate the poste- rior probabilities (PP) applying the majority rule consen- sus.
2.4 ZooBank Registration
The ZooBank registration number of the present work is: urn:lsid:zoobank.org:act:0D1078F7-850F-464B-B64B- AB64927AD677.
3 Results
3.1 Taxonomy
Order: Strombidiida Petz & Foissner, 1992
Family: Strombidiidae Fauré-Fremiet, 1970
Genus:Claparède & Lachmann, 1859
Species:sp. n.
3.2 Diagnosis
Cell size (40–60)×(20–28)μm,radish-like body with a bumped anterior end and an elongated tail-like posterior end, possessing a red eyespot in apical protru- sion. Buccal field about 1/3 of cell length. Extrusomes needle-shaped, about 10×0.5μm in size, tightly arranged along the girdle kinety. Single macronucleus spindle- shaped. Adoral zone of membranelles composed of 12–16 collar and 9–12 buccal membranelles.Girdle kinety located in the anterior 2/5 of cell, ostensibly continuous and composed of 45–60 dikinetids. Ventral kinety com- posed of 16–19 dikinetids, longitudinally extending from posterior 1/3 of cell to posterior end.
3.3 Type Locality and Habitat
Coastal waters off Songyu Wharf(118˚05΄E, 24˚45΄N), Fujian Province, China. Water samples collected on May 19, 2018 with salinity 22.4, temperature 26.5℃,and pH 7.9.
3.4 Deposition of Slides
A protargol slide (registration number HYY20180519-1) containing the holotype specimen and a slide (registra- tion number HYY20180519-2) containing the paratype specimens are deposited in the Laboratory of Protozool- ogy, Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China.
3.5 Etymology
Composite of the Greek prefix(false, being apparently like) and the species name, referring this species similar toin morphol- ogy.
3.6 Description
Cell size (40–60)×(20–28)μm, and about (36–54)×(18–25)μm after protargol impregnation (Table 1). Body shape generallyradish-like, with a bumped anterior end and an elongated tail-like posterior portion. Some thin individuals elongated conical shaped. The widest por- tion at anterior 1/3, posterior 2/3 tapering with a shar- pened end (Figs.1A, 1B, 2A, 2B, 2C). Anterior end with a prominent apical protrusion about 5μm high. Numerous spherical red pigment granules, each about 1–2μm across, densely concentrated at the apical protrusion, forming the ‘eyespot’ (Figs.1A, 1D, 2D, 2F, 2G, arrowheads).

Table 1 Morphometric data of Strombidium pseudorapulum sp. n.
Notes: All data based on randomly selected protargol-impregnated specimens. Measurements in μm. Mean, arithmetic mean;, number of individuals examined; SD, standard deviation.
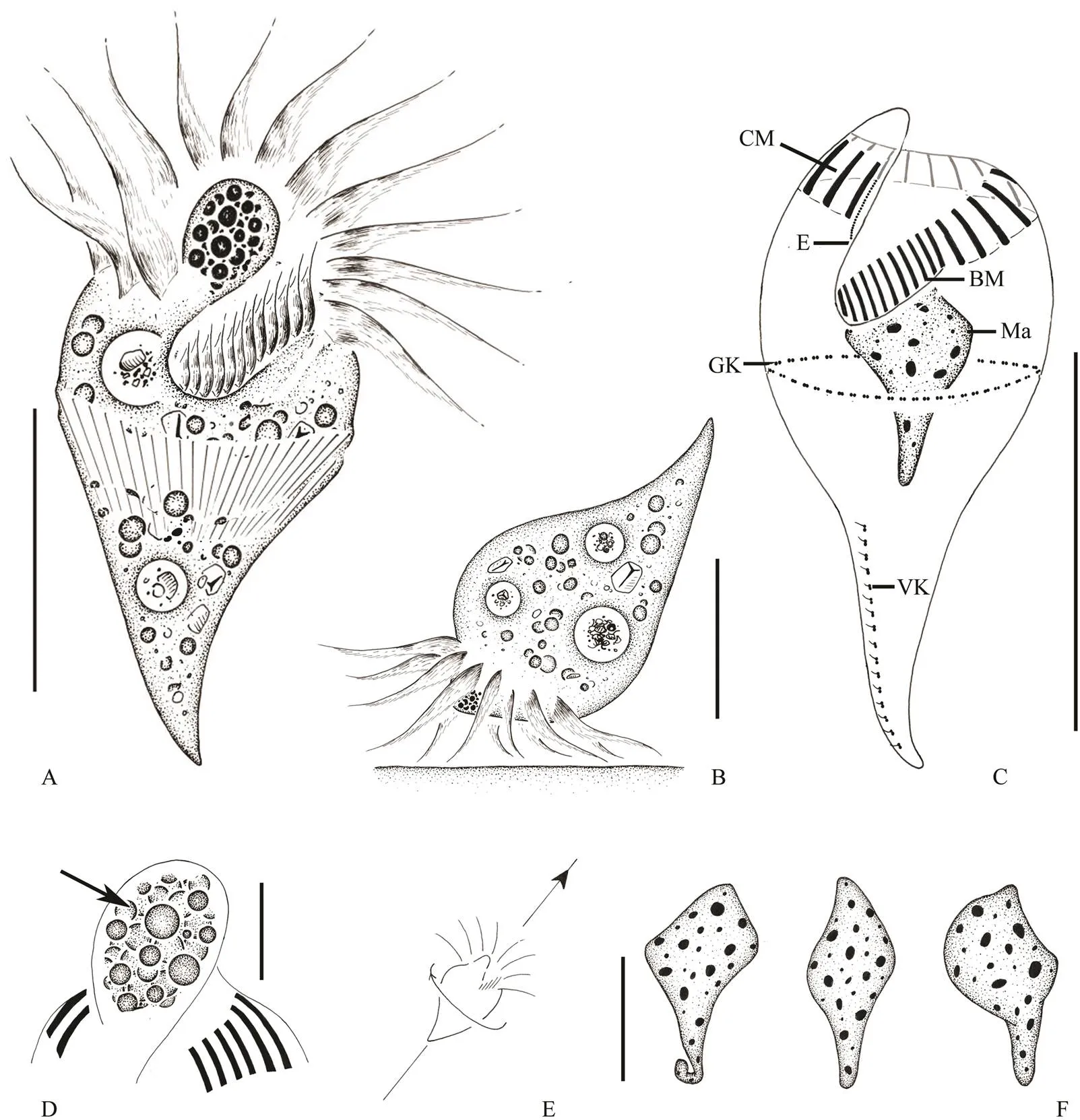
Fig.1 Strombidium pseudorapulum sp. n. from life (A, B, E) and after protargol impregnation (C, D, F). A, Ventral view of a representative specimen in vivo; B, Lateral view of a representative specimen when feeding; C, Infraciliature of both ventral and dorsal views; D, Arrow pointing to the details of ‘eyespot’ in apical protrusion; E, Swimming trace, arrowhead marking moving directions; F, Different shapes of macronucleus. BM, buccal membranelles; CM, collar membranelles; E, endoral membrane; GK, girdle kinety; Ma, macronucleus; VK, ventral kinety. Scale bars: A, 30µm; B, 20µm; C, 30µm; D, 5µm; F, 10µm.

Fig.2 Photomicrographs of Strombidium pseudorapulum sp. n. from live cells (A–H) and after protargol impregnation (I–M). A, B, Ventral (A) and lateral (B) views of radish-like individuals; C, Slim individual; D, Showing the apical pro- trusion (‘eyespot’, arrowhead) and the extrusomes (arrows); E, Showing the collar membranelles; F, G, To show the ‘eyespot’ (arrowheads); H, Apical view, to show the adoral zone of membranelles; I, Oral field, to show the buccal and collar membranelles; J, Showing the collar membranelles; K, Showing the macronucleus; L, Showing the girdle kinety; M, Oral primordium (arrow); N, The ventral kinety. BM, buccal membranelles; CM, collar membranelles; GK, girdle kinety; Ma, macronucleus; VK, ventral kinety. Scale bar: 30µm.
Hemitheca not observed. Cytoplasm colourless, con- taining lipid droplets 2–4μm across and food vacuoles about 7 μm in diameter with food particles, which render the yellow colour of cell under low magnifications (Figs.2A–D). Extrusomes needle-shaped, about 10×0.5μm in size, obliquely arranged in a row surround the middle body (Fig.1A). The anterior ends of extrusomes attached to cell surface about 5μm above girdle kinety (Figs.2D, L). Sin- gle macronucleus spindle-shaped, located in cell center, about (19–22)×(8–13)μmafter protargol impre- gnation (Fig.2K). Micronucleus and contractile vacuole not observed.
Locomotion special, usually with anterior end of cell attaching to substrate by adoral membranelles and poste- rior cell portion lifted (Fig.1B). Cell rapidly swim in a straight line by rotating cell axis after being stimulated (Fig.1E).
Buccal cavity wide, extending obliquely to about 1/3 of cell length. Adoral zone of membranelles continuous (Figs.1C, 2I, 2J). Collar portion composed of 12–16mem- branelles, bases about 8μm wide, cilia about 15μm long. Buccal portion composed of 9–12 membranelles, bases 4– 5μm wide, cilia 4μm long. Endoral membrane located on the right side of the inner wall of buccal lip, composed of a single row of kinetosomes (Fig.2I).
Somatic ciliature comprises a girdle kinety and a ven- tral kinety (Figs.1C, 2L, 2N). Girdle kinety horizontally positioned in anterior 2/5 of cell, closed and composed of 45–60 densely spaced dikinetids, each having a 2μm longcilium on the left basal body. Ventral kinety starting about 14 μm below the girdle kinety and extending longitude- nally along the ‘tail’ to the posterior end, occupying about the posterior 1/3 of cell. Ventral kinety is composed of 16–19 dikinetids andeach dikinetid possesses a cilium about 2μm long on its anterior basal body. The oral pri- mordium under thegirdle kinety and left of ventral ki- nety was observed in an individual cell with early divi- sion (Fig.2M).
3.7 SSU rRNA Gene Sequence and Phylogenetic Analyses
T he SSU rRNA gene sequence ofsp. n. was deposited in GenBank with acces- sion number MT274320. Its length is 1706bp and GC con- tent 46.25%. The Maximum likelihood (ML) and Baye- sian inference (BI) trees were nearly identical in their to- pology, thus they were combined in one tree with their support values marked in order (Fig.3). In both trees,sp. n. clusters withas sis- ter species (100% ML, 1.00 BI), then they cluster with,,,,, andforming a large clade with high support (90% ML, 1.00 BI). Most species of the genusgroup can be combined together as one of the main clades in the family Strom- bidiidae except for,,and, which nest within another clade composed of other strombidiid genera.
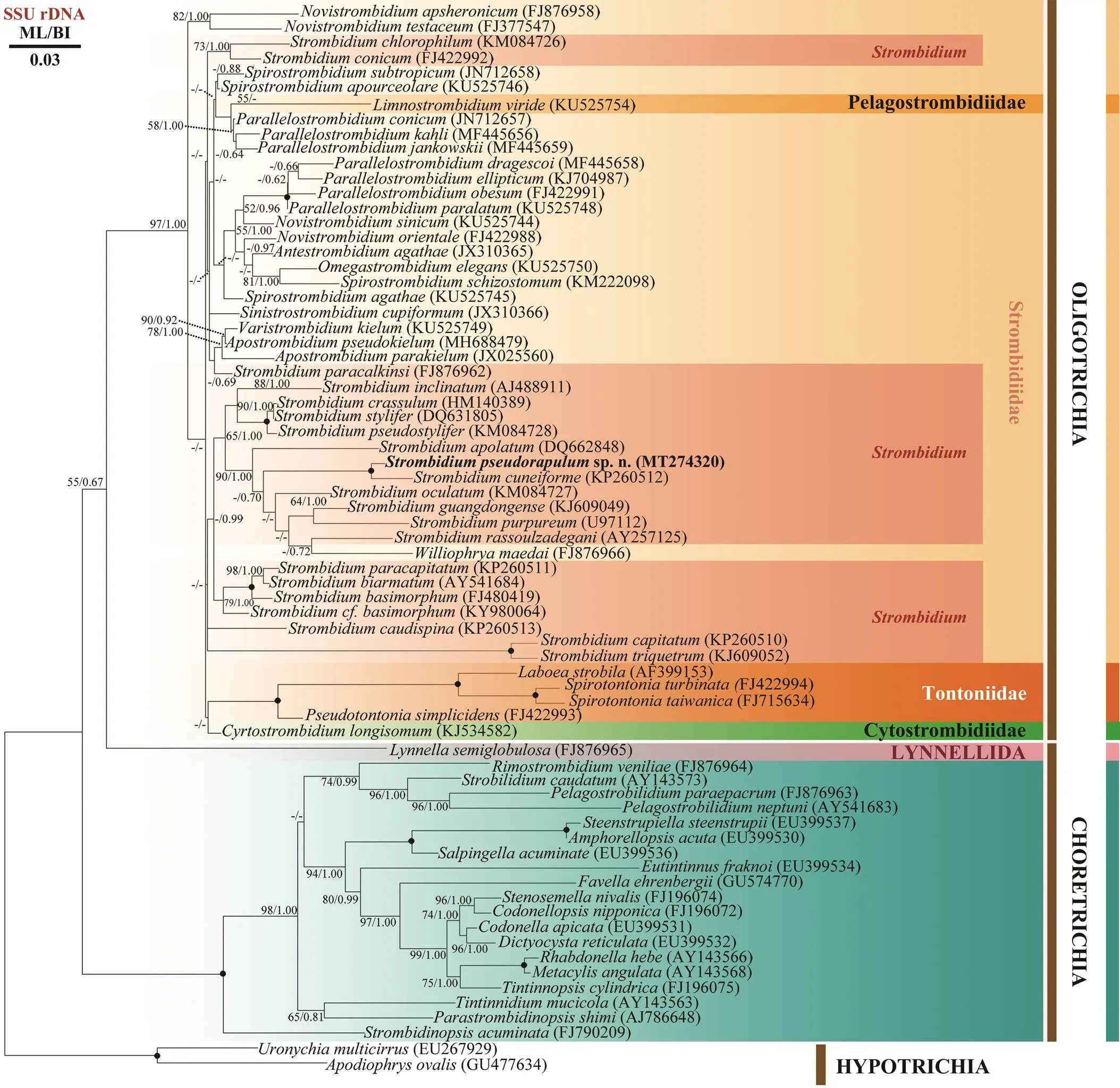
Fig.3 SSU rRNA gene phylogenetic tree of oligotrich and choreotrich ciliates with special focus on Strombidium spe- cies. Numbers at the nodes represent support values in the following order: Maximum Likelihood (ML) bootstrap val- ues/Bayesian inference (BI) posterior probabilities. Black solid dots indicate full support in all analyses (100% ML; 1.00 BI). Hyphens (-) reflects values that are below 50% or 0.50. Newly sequenced species is shown in bold font. All branches are drawn to scale. The scale bar corresponds to 3 substitutions per 100 nucleotide position.
4 Discussion
4.1 Comparison of Strombidium pseudorapulum sp. n. with Congeners
The most distinguishable features ofsp. n. are the presence of ‘eyespot’ in apical protrusionof cell and itsradish-like body shape with elongated ‘tail’. In terms of body shape, our new species is very similar to three congeners,.,Song, 2015,Song, 2015, and(Yagiu, 1933) Kahl, 1934 (Xu, 2006; Song, 2015a, b; Hu, 2019).
andsp. n. are nearly the same in their body shapes. However, it can be clearly distinguished from the new species by the following characters: absence of red ‘eyespot’ (. pres- ence),less buccal membranelles (7–9. 9–12), girdle kinety located posterior to equator of cell (girdle kinety in anterior 2/5 of cell), more dikinetids in ventral kinety (17–29. 16–19), and shape ofmacronucleus (ellipsoid- dal. spindle) (Song., 2015a).
Like, our new species also possesses typical ‘eyespot’ and similar locomotion pattern. How- ever, the former differs from the new species by the body shape (wedge. radish-like), more dikinetids in the ven- tral kinety (24–33. 16–19), and shape of macronu- cleus (ovoidal or elongate-ovoid. spindle) (Song., 2015b).
sp. n. can be identified fromwhich was reported from sea urchin by: free living lifestyle (. endocommensal), less buccal membra- nelles (9–12. 28–31), and more dikinetids in the ven- tral kinety (16–19. 5) (Xu, 2006).
The SSU rRNA gene sequence ofsp. n.shares about 98% similarity to that ofwith only 31 unmatched nucleotides. It differs fromby 102 unmatched nucleotides. Molecular data ofis not available so far.
Both morphological and molecular data support the va- lidity of this new species.
4.2 SSU rRNA-Based Phylogeny
In our phylogenetic analyses, the family Stombidiidae is generally composed of two large clades. One clade is composed of most species of, while the other clade contains other genera of strombidiids, which imply that these two clades probably represent different evolu- tionary processes as suggested in previous study (Liu, 2016).species are distributed among sev- eral different clades suggesting that this genus is not mo- nophyletic, although previous studies have strongly arguedfor its monophyly (Agatha and Strüder-Kypke, 2014; Tsai, 2015). In addition, our new speciessp. n. clusters withfirst as sister spe- cies, then forms a clade with,,,, and. Interestingly, all species within this clade were reported with pigment spot (‘eyespot’) in apical protru- sion of cell except for, the living morphol- ogy of which is incomplete so far.shares nearly the same body shape with the new species but has no ‘eyespot’. This confirms the viewpoint of previous study that the pigment spot might represent a synapo- morphy for this clade of strombidiids (Liu, 2016). Furthermore,has a similar taper poste- rior ‘tail’ as, but they do not cluster to- gether, suggesting that the ‘tail’ might be the result of in- dependent evolution inspecies.
Acknowledgements
This work was supported by the National Natural Sci- ence Foundation of China (No. 31761133001), the Guang- dong MEPP Fund (No. GDOE (2019) A23), the Key Spe- cial Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0405), and Innovation Academy of South China Sea Ecology and Environmen- tal Engineering, Chinese Academy of Sciences (No. ISEE 2018PY01).
Adl, S. M., Bass, D., Lane, C. E., Lukeš, J., Schoch, C. L., Smir- nov, A., Agatha, S., C., Berney, Brown, M. W., Burki, F., Cár-denas, P., Čepička, I., Chistyakova, L., del Campo, J., Dun- thorn, M., Edvardsen, B., Eglit, Y., Guillou, L., Hampl, V., Heiss, A. A., Hoppenrath, M., James, T. Y., Karnkowska, A., Karpov,S., Kim, E., Kolisko, M., Kudryavtsev, A., Lahr, D. J. G., Lara, E., Le Gall, L., Lynn, D. H., Mann, D. G., Massana,R., Mit- chell, E. A. D., Morrow, C., Park, J. S., Pawlowski, J. W., Po- well, M. J., Richter, D. J., Rueckert, S., Shadwick, L., Shi- mano, S., Spiegel, F. W., Torruella, G., Youssef, N., Zlato- gursky, V., and Zhang, Q., 2019. Revisions to the classifi- cation, nomenclature, and diversity of eukaryotes., 66 (1): 4-119.
Agatha, S., 2011. Global diversity of aloricate Oligotrichea (Pro-tista, Ciliophora, Spirotricha) in marine and brackish sea wa- ter., 6 (8): e22466.
Agatha, S., and Riedel-Lorjé, J.C., 2006. Redescription ofDaday, 1887 (Ciliophora: Spirotricha) andunification of tintinnid terminology.,45: 137-151.
Agatha, S., and Strüder-Kypke, M. C., 2014. What morphology and molecules tell us about the evolution of Oligotrichea (Alveolata, Ciliophora)., 53 (1): 77.
Agatha, S., Strüder-Kypke, M. C., Beran, A., and Lynn, D. H., 2005.(Montagnes and Taylor, 1994) andnov. spec. (Ciliophora, Oligotri- chea): Phylogenetic position inferred from morphology, onto- genesis, and gene sequence data., 41 (1): 65-83.
Claparède, E., and Lachmann, J., 1859. Études sur les infusoires et les rhizopodes.,6: 261-482.
Elangovan, S. S., and Gauns, M., 2017. New species of ciliates (Genus:sp.) from hypoxic waters of the Bay of Bengal, northern Indian Ocean.,46 (4): 686-691.
Gao, F., Warren, A., Zhang, Q., Gong, J., Miao, M., Sun, P., Xu, D. P., Huang, J., Yi, Z. Z., and Song, W. B., 2016. The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata)., 6: 24874.
Gao, S., Gong, J., Lynn, D., Lin, X. F., and Song, W. B., 2009. An updated phylogeny of oligotrich and choreotrich ciliates (Protozoa, Ciliophora, Spirotrichea) with representative taxa collected from Chinese coastal waters., 7 (2): 235-242.
Hall, T. A., 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at:. In- formation Retrieval Ltd., London, c1979-c2000.
Hu, X. Z., Lin, X. F., and Song, W. B., 2019.. Science Press, Beijing, 416-429.
Kim, Y. O., Kim, S. Y., Lee, W. J., and Choi, J. K., 2010. New observations on the choreotrich ciliateFauré-Fremiet 1924, and comparison withJeong., 2004., 57 (1): 48-55.
Li, J. M., Liu, W. W., Gao, S., Warren, A., and Song, W. B., 2013. Multigene-based analyses of the phylogenetic evolu- tion of oligotrich ciliates, with consideration of the internal transcribed spacer 2 secondary structure of three systemati- cally ambiguous genera., 12 (3): 430-437.
Liu, W. W., Jiang, J. M., Xu, Y., Pan, X. M., Qu, Z. S., Luo, X. T., El-Serehy, H. A., Warren, A., Ma, H. G., and Pan, H. B., 2017. Diversity of free-living marine ciliates (Alveolata, Ci- liophora): Faunal studies in coastal waters of China during the years 2011–2016., 61: 424-438.
Liu, W. W., Xu, D. P., Ma, H. G., Al-Farraj, S. A., Warren, A., and Yi, Z. Z., 2016. Taxonomy and molecular systematics of three oligotrich (sl) ciliates including descriptions of two new species,sp. nov. andsp. nov. (Protozoa, Ciliophora)., 14 (5): 452-465.
Liu, W. W., Yi, Z. Z., Li, J. Q., Warren, A., Al-Farraj, S. A., and Lin, X. F., 2013. Taxonomy, morphology and phylogeny of three new oligotrich ciliates (Protozoa, Ciliophora, Oligotri- chia) from southern China., 63 (12): 4805-4817.
Liu, W. W., Yi, Z. Z., Warren, A., Al-Rasheid, K. A., Al-Farraj, S. A., Lin, X. F., and Song, W. B., 2011. Taxonomy, mor-phology and molecular systematics of a new oligotrich ciliate,gen. nov., sp. nov., with redescriptions ofand(Protozoa, Ciliophora, Oligotrichia)., 9 (3): 247-258.
Liu, W. W., Yi, Z. Z., Xu, D. P., Clamp, J. C., Li, J. Q., Lin, X. F., and Song, W. B., 2015. Two new genera of planktonic ciliates and insights into the evolution of the family Strom- bidiidae (Protista, Ciliophora,Oligotrichia)., 10(6): e0131726.
Liu, W. W., Zhang, K., Chen, C., Li,J. M., Tan, Y., Warren, A., Lin, X. F., and Song, W., 2019. Overview of the biodiversity and geographic distribution of aloricate oligotrich ciliates (Pro- tozoa, Ciliophora, Spirotrichea) in coastal waters of southern China., 17 (8): 787-800.
Lynn, D., 2008.. Springer Science and Business Media, Dordrecht, 605pp.
McManus, G. B., Xu, D. P., Costas, B. A., and Katz, L. A., 2010. Genetic identities of Cryptic species in thecluster, including a description ofn. sp., 57 (4): 369-378.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L., 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions., 71 (2): 491-499.
Miller, M. A., Pfeiffer, W., and Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylo- genetic trees.. New Orleans, LA, 1-8.
Nylander, J., 2004. MrModeltest v2. 2 Uppsala: Evolutionary Biology Centre, Uppsala University.
Ronquist, F., and Huelsenbeck, J. P., 2003. MrBayes 3: Baye- sian phylogenetic inference under mixed models., 19 (12): 1572-1574.
Song, W. B., 2005. Taxonomic description of two new marine oligotrichous ciliates (Protozoa, Ciliophora)., 39 (3): 241-252.
Song, W. B., Warren, A., and Hu, X. Z., 2009.. Science Press, Beijing, 307-353.
Song, W., Li, J. M., Huang, Y. Y., Hu, X. Z., Liu, W. W., Al-Rasheid, K. A., and Miao, M., 2018. Morphology of three aloricate choreotrich ciliates, including description of a new speciessp. n. (Ciliophora, Cho- reotrichia), and phylogeny of the genus., 57: 153-167.
Song, W., Li, J. M., Liu, W. W., Al-Rasheid, K. A., Hu, X. Z., and Lin, X. F., 2015a. Taxonomy and molecular phylogeny of fourspecies, including description ofsp. nov. (Ciliophora, Oligotrichia)., 13 (1): 76-92.
Song, W., Zhao, X., Liu, W. W., Hu, X. Z., Al-Farrajc, S. A., Al-Rasheid, K. A. S., Song, W. B., and Warren, A., 2015b. Biodiversity of oligotrich ciliates in the South China Sea: Description of three newspecies (Protozoa, Ciliophora, Oligotrichia) with phylogenetic analyses., 13 (6): 608-623.
Stamatakis, A., 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mix- ed models., 22 (21): 2688-2690.
Stamatakis, A., Hoover, P., and Rougemont, J., 2008. A rapid bootstrap algorithm for the RAxML web servers., 57 (5): 758-771.
Tsai, S. F., Chen, W. T., and Chiang, K. P., 2015. Phylogenetic position of the genus, with a description ofnov. spec. and a re- description ofLynn and Gil- ron, 1993 (Protozoa, Ciliophora) based on live observation, protargol impregnation, and 18S rDNA sequences., 62 (2): 239-248.
Wilbert, N., 1975. Eine verbesserte technik der protargo-lim- pragnation fur ciliaten., 64: 171-179.
Wilbert, N., and Song, W. B., 2005. New contributions to the marine benthic ciliates from the Antarctic area, including des-cription of seven new species (Protozoa, Ciliophora)., 39 (13): 935-973.
Xu, D. P., Song, W. B., Sun, P., and Chen, X. R., 2006. Mor- phology and infraciliature of the oligotrich ciliate(Yagiu, 1933) Kahl, 1934 (Protozoa, Ciliopho- ra, Oligotrichida) from the intestine of sea urchinAgassiz., 1113 (1): 33-40.
Xu, D. P., Sun, P., Song, W. B., and Warren, A., 2008. Studies on a new endocommensal ciliate,nov. sp. (Ciliophora, Oligotrichida), from the intestine of the sea urchin(Camarodonta, Echino- ida)., 23: 273-278.
. E-mail: wwliu@scsio.ac.cn
E-mail: linxf@xmu.edu.cn
December 11, 2019;
December 17, 2019;
March 31, 2020
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Dynamic Diurnal Changes in Green Algae Biomass in the Southern Yellow Sea Based on GOCI Images
- Nutrient Enrichment Regulates the Growth and Physiological Responses of Saccharina japonica to Ocean Acidification
- Isolation of Enterococcus faecium with Feeding Attractant Function from Pacific White Shrimp (Litopenaeus vannamei) Intestine
- Transcriptomic Profiling of the Immune Response to Crowding Stress in Juvenile Turbot (Scophthalmus maximus)
- Reducing the Common Environmental Effect on Litopenaeus vannamei Body Weight by Rearing Communally at Early Developmental Stages and Using a Reconstructed Pedigree
- Can Langmuir Circulations Solve the Problem of Insufficient Upper-Ocean Mixing?
