Reducing the Common Environmental Effect on Litopenaeus vannamei Body Weight by Rearing Communally at Early Developmental Stages and Using a Reconstructed Pedigree
2020-09-28KONGZhangweiKONGJieHAODongchunLUXiaJIANTanMENGXianhongLUOKunCAOBaoxiangSUIJuanLIXupengandLUANSheng
KONG Zhangwei, KONG Jie, HAO Dongchun, LU Xia, JIAN Tan, MENG Xianhong, LUO Kun, CAO Baoxiang, SUI Juan, LI Xupeng, and LUAN Sheng, *
Reducing the Common Environmental Effect onBody Weight by Rearing Communally at Early Developmental Stages and Using a Reconstructed Pedigree
KONG Zhangwei1), 3), KONG Jie1), 2), HAO Dongchun1), LU Xia1), 2), JIAN Tan1), 2), MENG Xianhong1), 2), LUO Kun1), 2), CAO Baoxiang1), 2), SUI Juan1), 2), LI Xupeng1), 2), and LUAN Sheng1), 2), *
1),,,266071,2),,266235,3),,266003,
Heritability estimates may be severely biased when a large common environmental effect on a family arises from a long- lasting separate rearing at early stages (SRES) in traditional selective breeding programs, especially when bred populations have weak genetic ties. Communal rearing at early stages (CRES) may reduce common environmental effect since all families are reared in the same environment immediately after hatching. Here, we compared the effects of CRES and SRES strategies on genetic parameter estimation for harvest body weight in a selective breeding populationwith a small number of half-sib families. Genetic parameters of each strategy were estimated by using animal models excluding and including the common environmental effect (Model 1 and Model 2, respectively). Heritability estimates for body weight were 0.21±0.06 (<0.05) and 0.69±0.09 (<0.05) for CRES and SRES, respectively, in Model 1, and 0.21±0.06 (<0.05) and 0.52±0.27 (>0.05) in Model 2. The ratio of common environmental variance to phenotypic variance was 0.002±0.000 and 0.071±0.112 for CRES and SRES, respectively. Neither strategy precisely partitioned the common environmental variance according to likelihood ratio test. Lower heritability for body weight in CRES than in SRES implied that a large common environmental variance was confounded with additive genetic variance and was not effectively partitioned in SRES. Moreover, genetic correlation of body weight between the two strategies was 0.75±0.15, indicating that family rankings truly changed. The CRES should be followed in the selective breeding program of shrimp, especially in a population with a shallow pedigree and weak genetic ties between families.
common environmental effect;heritability;; parentage assignment
1 Introduction
Pacific white shrimp,, accounts for approximately 60% of global shrimp aquaculture pro- duction (FAO, 2018). In last 20 years, the benefits of selective breeding of shrimp have been clearly demonstrated. For example, such program has produced considerable ge- netic gains of growth traits (4.4%–21.0%), disease resistance (1.7%–6.3% for white spot syndrome virus; 12.4%– 18.4% for Taura syndrome virus), and survival (1.1%– 5.7%) (Fjalestad., 1997; Argue,., 2002; Gitterle., 2007; Andriantahina,., 2012; Luan., 2012; Huang., 2012).
Genetic improvement programs for shrimp often use family and sib-selection strategies. These strategies require to use physical tags, such as visible implant elasto-mers (VIE), to identify specific families and individuals within a family for data collection and pedigrees tracking (Godin., 1996; Jerry., 2001; Arce., 2003). With these strategies, families must be separately reared for 60–90 days until juvenile shrimp are large enough to be physically tagged. The variance of the full-sib group effect, which is also called common environmental variance, becomes confounded with additive genetic variance if not properly partitioned in statistical models (Ninh., 2011). For a breeding population with many half-sib fa- milies, it is feasible to robustly partition this common en- vironmental variance due to strong genetic ties among fa- milies. However, this task becomes more challenging when the bred population includes only a few half-sib families, which occurs when trying to selectively breed shrimp due to unsuccessful nested mating (Kenway., 2006; Sui., 2015).
To resolve this problem, highly polymorphic microsatellites for parentage inference can be employed. Parentage assignment using molecular markers allows families to remain together after hatching, thereby substantiallyreducing the common environmental effects (Rodzen., 2004), making heritability estimates more accurate and ensuring a higher selection response (Fjalestad., 2003). Pedigree analysis using molecular markers to estimate ge- netic parameters has already been applied to various fish species, such as Atlantic salmon (Norris., 2000), white sturgeon (Rodzen., 2004), and common carp (Ninh., 2013). Similarly, forgenetic markers have been used to characterize genetic diversity for individual identification and assign parentage (Cruz., 2004; Valles-Jimenez., 2004; Garcia., 2007; Pérez- Enriquez., 2016). However, few studies have focus- ed on reducing the common environmental effect on the shrimp growth traits through communal rearing early in selective breeding programs (Nolasco-Alzaga., 2017).
We designed a communal rearing at early stages (CRES) strategy to reduce the common environmental effect in the breeding program ofwith a small number of half-sib families. The pedigree of all tested individuals was reconstructed by assigning parentage using microsatellite markers. For comparison, we also used a separate rearing at early stages (SRES) strategy to a shrimp population with a pedigree structure similar to that of CRES group to estimate the genetic parameters. We determined the gene- tic and phenotypic associations between two groups to ver- ify the existence of a significant genotype by environment interaction (G×E). The primary objective of this study was to test the applicability of CRES for heritability estimation of a special shrimp population with a weak genetic tie among families.
2 Methods
2.1 Base Population of Shrimp
broodstock population came from a breed- ing program carrying out at Marine Genetic Breeding Cen- ter of Ministry of Agriculture, Yellow Sea Fisheries Research Institute (Qingdao, China). This breeding program was established from seven batches ofbrood- stock introduced from America and Singapore from June to July, 2011. The base population (G0) was produced us- ing an incomplete diallel cross experiment (Sui., 2015), and a nested mating design was used to produce all subsequent generations. This shrimp is neither an endangered nor a protected species, and all experiments described here complied with the Law of the People’s Republic of China on the Protection of Wildlife (http://www.china.org. cn/english/environment/34349.htm).
2.2 Production of Shrimp Families
The breeding candidates came from G4 generation, and were chosen based on a gross observation of reproductive maturity. Females displayed orange-colored ovaries occu- pied by a large area of the cephalothorax, and males had a full and white spermatophore. To identify different indivi- duals, each broodstock was tagged with a unique, alpha- numeric eyestalk ring. Full-sib and half-sib families were produced by artificial insemination following a nested mating design. Simultaneously, paternal and maternal half- sib families were produced by mature breeders. A total of 142 sires and 136 dams were selected from G4 generation to produce 155 full-sib families named as G5. Each fertilized female was transferred to a separate 170-L white tank and kept for 12h until her eggs began to hatch.
2.3 Rearing and Testing of Families
The hatched larvae underwent through six nauplii-stages, three zoea-stages, and three mysis-stages in 12 days before they metamorphosed into post-larvae. Shrimp were grow out following a standard commercial practice as re- cently described (Lu., 2016). One hundred full-sib families out of 155 in total with similar average body lengths were randomly selected and reared using CRES and SRES strategies.
2.3.1 Communal rearing at early stages (CRES)
On day 20 post-hatching, a total of 5000 post-larvae (PL), 50 each family, were stocked in a 3m3tank. For each individual, the stocking body length was measured. At day 50 post-hatching, all of them were moved to a larger tank (20m3). From day 80 post-hatching, the juveniles were cultured in 100m3tanks. Four times a day, the shrimp were fed with a commercial dry feed containing 12% moisture, 42% crude protein and 17% crude ash at 3%–5% of live weight. The tank water was changed from 10% to 30% of the total according to key water quality parameters (.., pH, dissolved oxygen, and total ammonia). On day 120 post- hatching, the wet weight (g) of individuals was measured, and a small piece of muscle tissue was isolated from each shrimp and preserved in 95% ethanol for DNA extraction.
2.3.2 Separate rearing at early stages (SRES)
On day 20 post-hatching, PL from 100 families, 1500 each, were transferred to separate 170L tanks, one each family, and cultured continually. On day 50 post-hatching, 700 post-larvae each family were randomly selected and transferred to separate larger tanks (3m3). On day 100 post-hatching, when the mean body weight of juveniles reached 3–4g, 35 shrimp were randomly selected each family, 76 in total, and individually tagged by injecting visible implant elastomer (VIE) at five abdominal positions. A unique combination of VIE colors (green, blue, orange and red) was used to identify families. The tagged shrimp were transferred to a 1m×5.6m×5.6m tank, and reared as were done in standard management practices. The same commercial feed, containing 12% moisture, 42% crude protein and 17% crude ash, was used to feed the shrimp. The feeding rate was adjusted according to live weight of shrimp, which was similar with that used in CRES strategy. The tank water was changed from 15% to 30% of the total in volume depending on the shrimp growth stage. Finally, on day 177 after hatching, the wet weight (g) of individuals was measured.
To reduce difference in average harvested body weight of shrimp between CRES and SRES, the rearing period for CRES was reduced due to its higher growth rate com- pared to that of SRES.
2.4 DNA Extraction and Microsatellite Analysis
Genomic DNA was obtained from the muscle tissues of the parents (174 individuals, the parents of 100 families, including 92 sires and 82 dams) and their progeny (1451 individuals) using a phenol/chloroform extraction method as described previously (Liu., 2000). Genomic DNA was then resuspended in TE buffer (10mmolL−1Tris-HCl, pH 7.6, 0.1mmolL−1EDTA) at a final concentration of 100ngμL−1.
Thirteen polymorphic microsatellites were selected for genotyping. Of them, 12 were polymorphic, which were optimized into three multiplex PCR sets, TM6, TUMXLv3.1, TUMXLv7.121, TUMXLv9.90 and 1103; Lv12, Lv14, TUMXLv10.33 and 10592b; TUMXLv8.256, TUMXLv 8.220 and TUMXLv7.56), respectively (Li., 2016), and a single PCR, TUMXLv10.14a. Amplification was per- formed in a 25-μL volume containing 100ng DNA, 0.5mmolL−1dNTP (each), 25mmolL−1MgCl2, 2.5μL of 10×buffer, ddH2O appropriate for each microsatellite set, 1μL ofpolymerase (AB Gene) and 2μmol specific pri- mers (each). Amplification was performed in a Gene Amp PCR System, 9600 Thermocycler, following the protocol described by Li. (2016). The products of PCR were tested using agarose gel electrophoresis. Subsequently, 1.0μL of the PCR product mixture was added to 10μL frag- ment size standard labeled with the red fluorescent dye 6- carboxy-χ-rhodamine (ROX) (Brondani and Grattapaglia, 2001). Microsatellite alleles were separated by capillary electrophoresis using an ABI 3100 DNA analyzer (App- lied Biosystems). Genotypes were then read using Gene- Mapper v4.0 software (Applied Biosystems, Foster City, California, USA).
2.5 Data Analysis
2.5.1 Assigning parentage
Parentage assignment was performed through a likeli- hood-based approach with CERVUS v3.0 software (Ka- linowski., 2007). The simulation analysis applied the following parameters: 1) 100% of candidate parents sam- pled, and 2) 95% genotyped at a default type I error rate of 1%. Furthermore, <4 mismatched alleles were permit- ted for offspring and their given parents, and an unambi- guous pedigree was generated, in which only those as- signments with ≥95% confidences were accepted.
2.5.2 Variance components and heritability estimates
To calculate the variance components of shrimp body weight at harvest, the average information REML method was used in ASReml (Gilmour., 2009). Two different animal models were implemented:

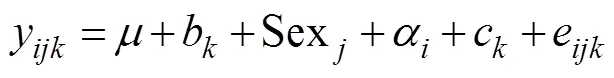

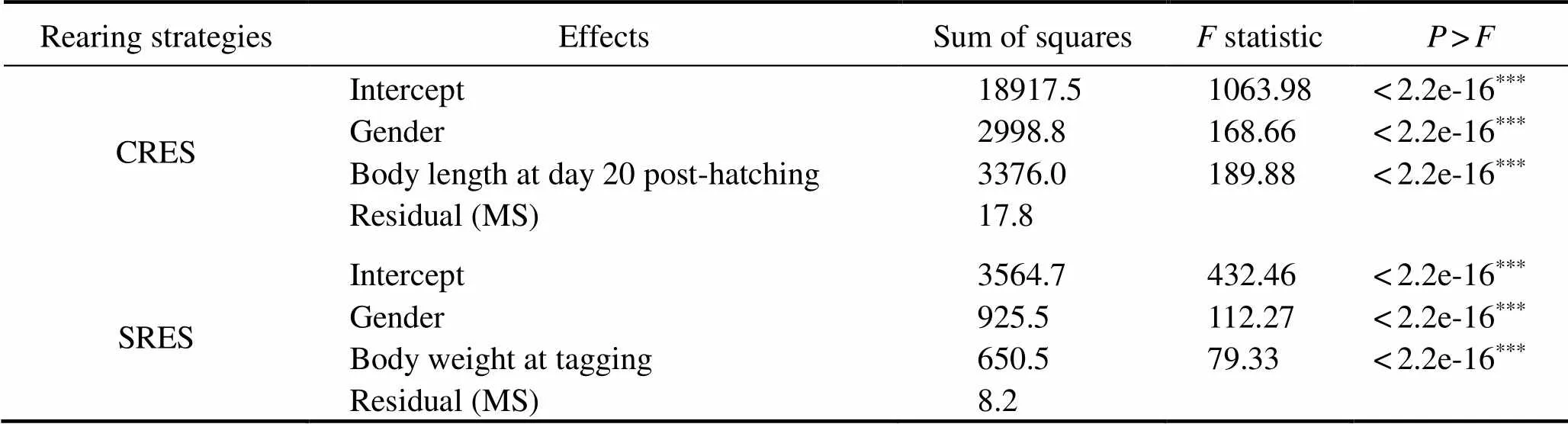
Table 1 Analysis of variance of harvest body weight of L. vannamei in CRES and SRES strategies:tests of the fixed effects using Wald test
Notes:***<0.0001; CRES, communal rearing at early stages; SRES, separate rearing at early stages.
The likelihood ratio test (LRT) was applied to body weight to determine the statistical significance of the ran- dom effect of interest (Gilmour., 2009; Wilson., 2010). Specifically, the models with or without the random effect were compared using LRT as follows:

where ln1and ln2are the natural logarithms of likeli- hood from Model 1 without the common environmental effect and Model 2 with the common environmental ef- fect, respectively. The asymptotic distribution of the like- lihood ratio follows the chi-square (2) distribution with 1 degree of freedom (Stram., 1994; Sae-Lim., 2013).
Genetic correlations of body weight between the CRES and SRES strategies were estimated with bivariate animal models. The additive genetic variance-covariance matrix was calculated following this form:

To test whether the heritability estimates for body weight were significantly different from zero, and whether the ge-netic correlation between the two groups differed signi- ficantly from zero, we relied on the-score test (Nguyen., 2007):

whereXis the estimate of heritability or genetic correlation, andσis its corresponding standard error. Here, bothXandσwere set to zero. The resulting-score was then tested against a large sample with a normal distribution.
3 Results
3.1 Descriptive Statistics
For CRES group, a total of 1451 offspring and 174 pa- rents were available for parentage assignment; of them, 1356 (93.4%) offspring were unambiguously assigned to 72 full-sib families including 18 half-sib ones (Table 2).
Descriptive statistics for harvest body weight in CRES and SRES are given in Table 2. There was fewer male off- spring than female offspring in CRES and SRES groups. This did not represent a lower survival of male than that of female because the gender of stocking post-larvae was un- known. Two genders differed slightly in their mean body weight in CRES and SRES. The coefficient of variation for harvest body weight for the whole population (CV=44%) of CRES was almost one-third larger than that of SRES (33%).

Table 2 Descriptive statistics for harvest body weight in CRES and SRES for harvest L. vannamei body weight
Notes:†The gender of 117 and 19 individuals cannot be ascertained due to small body sizes.††Refers to all individuals (male, female, and no-gender) that had a phenotype value. CRES, communal rearing at early stages; SRES, separate rearing at early stages;, number of;f,h, number of full-sib and half-sib families, respectively; SD, standard deviation; CV, coefficient variation.
Fig.1 shows that offspring survival for each family in CRES and SRES is generally not high. CRES had a great- er variance than SRES in the proportion of surviving fa- milies despite undergoing similar management techniques at all culture stages. The main reason for this result is like-ly that the original weight of individuals was lower in CRES than in SRES.
Pearson correlations revealed medium associations of stocking body length (tagging body weight) with harvest body weight of families in CRES (0.53±0.02) and SRES (0.42±0.11) groups. This indicated that stocking body length (tagging body weight) should be considered as co- variates in Models 1 and 2.
3.2 Variance Components and Genetic Parameters
Table 3 summarizes the obtained estimates of variance components, heritability, and2for shrimp growth traits using CRES and SRES. No significant difference was found between CRES and SRES in terms of phenotypic vari- ance (=1.13,0.01(1355,1226)=1.14,>0.01). For Model 1, the heritability estimate for body weight was moderate (0.21) in CRES, and was significantly different from zero (Z=3.56,<0.01); however, high heritability (0.68;=7.64,<0.01) was obtained in SRES. For Model 2, the heritability estimate for body weight was likewise moder- ate (0.21) in CRES and significantly different from zero (=3.62,<0.01), for which a low and non-significant2for body weight was observed (0.002;=20223,0.01; Table 3). In SRES, the heritability for body weight was still high (2=0.51±0.27,=1.89,>0.01) with a2value of 0.07±0.11 that was not significantly different from zero (=0.63,>0.01). The LRT suggested that common en- vironmental effect should not be included in these models, and it was non-significant for both shrimp-rearing strategies (CRES:2=0.0014,>0.05; SRES:2=0.251,>0.05).

Fig.1 The proportion of surviving L. vannamei for each fa-mily in CRES and SRES strategies. CRES, communal rear- ing at early stages; SRES, separate rearing at early stages.
The mean genetic correlation of harvest body weight between CRES and SRES was 0.75±0.15 (Table 3), significantly different from 1 (=1.67,<0.05, one-tailed test), suggesting a medium re-ranking effect on the estimated breeding value between two strategies.

Table 3 Estimates of variance components for L. vannamei in CRES and SRES strategies from two fitted mixed models (1 and 2 refer to Materials and Methods)
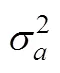
4 Discussion
4.1 Parental Assignment
Microsatellites are proven effective for accurately re- constructing the pedigree of many aquatic species (Kocour., 2007; Gheyas., 2009). Our proportion of success- ful assignments (93.4%) was similar to those reported in other aquatic species. For example, Dong. (2006) app- lied five microsatellites into ex- clusively assign 215 offspring to their parental pairs with 90.7% of success. Similarly, among strains of the giant freshwater prawn, a 90% cor- rect assignment of individuals was obtained using seven microsatellites (Karakete., 2011). In the study of Ngu- yen. (2014), the feasibility of pedigree reconstruction in aselection breeding program was starkly shown, wherein 10 microsatellites cor- rectly assigned 98% of parentages. Of course, the reliability and accuracy of inferring these kin relationships for parentage assignment can be affected in some cases. One crucial factor is the informativeness of microsatellites. The number and polymorphisms and null allele frequency of microsatellites, the number of parents, increased homozygosity in selective breeding and genotyping errors, determine the informativeness of microsatellites (Navarro., 2009; Kristjánsson., 2016). The medium-to- high success of parentage assignment in our study can be attributed to the larger number of potential families and parents, as well as lessness of microsatellites. Nevertheless, artificial genotyping errors were difficult to avoid, leaving some individuals imprecisely assigned to their true full-sib families. To improve the success of our parentage assignments, more markers are required.
4.2 Estimates of Heritability and Common Environmental Effect
In our study, a higher heritability for body weight was obtained in SRES compared with CRES, indicating that common environmental variance was not effectively partitioned from additive genetic one. Tan. (2016) suggested that inflating heritability forsurvival may arise when its additive and non-additive genetic effects are confounded. High heritability (0.32–0.42) for body weight inhas also been reported recently (Li., 2015; Sui., 2015), further highlighting that com- mon environmental variance of this economically valuable shrimp cannot be reliably estimated.
The lack or loss of genetic ties among families may lead to an inability to partition the common environmental va- riance from the phenotypic variance in some selective breeding programs (Castillo-Juárez., 2007). Using a large number of paternal half-sib families may improve the accuracy of quantifying the additive genetic effect in SRES strategy (Gitterle., 2005). Additionally, a thorough pedigree structure is elusive and time-consuming due to low rates of mating success and family survival, especiallyin selective breeding programs of, which fur- ther depends on broodstock maturity and artificial inse- mination technology.
Minimizing potential confounding of genetic and envi- ronmental variances is always a challenge in studying po- pulations with weak genetic ties (Jerry., 2004). In our study, applying CRES strategy notably reduced the com- mon environmental variance and improved the accuracy of our heritability estimate. The results are consistent with those of other researches to date on different aquaculture species like(Fishback., 2002),(Nguyen., 2014), andL. (Vandeputte., 2004, 2008; Kocour., 2007). For the last species, the response to selection for growth performance improved at a faster rate under CRES than under SRES strategy (Ninh., 2011).
4.3 Genetic Correlation
Though we had maintained similar rearing conditions between CRES and SRES strategies as close as possible, the genetic correlation between two strategies obtained us- ing Model 2 (0.75) was far below 1. This implied that a common environmental variance was not effectively partitioned from additive genetic variance, which led to a biased estimated breeding value for SRES group.
In addition, a re-ranking effect of the estimated breeding value for harvest body weight was detected among fa- milies in two strategies. In many developing countries, CRES strategy better approximates how shrimp are actually ma- naged and cultured than SRES strategy that is often used in shrimp selective breeding programs. Our study suggest- ed that SRES likely fosters adverse difference between breed- ing and production links. We propose that matching the breeding environment to its production environment in a better method is advantageous; hence, CRES strategy rather than SREC strategy is recommended for shrimp selective breeding programs.
5 Conclusions
To our best knowledge, this study is the first one to investigate the impact of CRES strategy on estimating the heritability and common environmental effect in apopulation with a small number of half-sib families by using parentage assignment technique. With CRES, a pedigree can be reconstructed using parentage assignment method with suitable microsatellites. Compared with SRES strategy, CRES possesses an obvious advantage in accurately estimating the heritability of body weight in; it reduces common environmental effect arising from separate rearing of families. Genetic correlation of body weight between two strategies revealed that changed ranks of families when estimated breeding values were confounded with common environmental effect in SRES. In a nutshell, CRES strategy with a short period of family- separated rearing was more effective in accurately estimating genetic parameters than SRES strategy did in this particularselective breeding program.
Acknowledgements
This research was financially supported by the National Key R&D Program of China (No. 2018YFD0901301), theShandong Province’s Agricultural Seed Improvement Pro- ject (No. 2017LZN011), the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 20603022020003), the China Agriculture Research System (CARS- 48), the Projects of International Exchange and Cooperation in Agriculture of Ministry of Agriculture and Rural Affairs of China–Science, Technology and Innovation Co- operation in Aquaculture with Tropical Countries, the In- troduction of International Advanced Agricultural Science and Technology Plan of China (No. 2016-X39), and the Ma- jor Applied Technology Innovation Project of Agriculture in Shandong Province (No. SD2019YY001).
Andriantahina, F., Liu, X., Huang, H., and Xiang, J., 2012. Response to selection, heritability and genetic correlations between body weight and body size in Pacific white shrimp,., 30: 200-205, http://dx.doi.org/10.1007/s00343-012- 1066-2.
Arce, S. M., Argue, B. J., Thompson, B. A., and Moss, S. M., 2003. Evaluation of a fluorescent, alphanumeric tagging system for penaeid shrimp and its application in selective breeding programs., 121: 313-326, https://doi.org/10. 1016/S0044-8486(03)00267-9.
Argue, B. J., Arce, S. M., Lotz, J. M., and Moss, S. M., 2002. Se- lective breeding of Pacific white shrimp () for growth and resistance to Taura syndrome virus., 204: 447-460, https://doi.org/10.1016/S0044-8486(01) 00830-4.
Castillo-Juárez, H., Casares, J. C. Q., Campos-Montes, G., Ville- la, C. C., Ortega, A. M., and Montaldo, H. H., 2007. Heritabi- lity for body weight at harvest size in the Pacific white shrimp,(), from a multi-environment experiment using univariate and multivariate animal models., 273: 42-49, https://doi.org/10.1016/j.aquaculture. 2007.09.023.
Cruz, P., Ibarra, A. M., Mejia-Ruiz, H., and Gaffney, P. M., 2004. Genetic variability assessed by microsatellites in a breeding pro- gram of Pacific white shrimp ().,6: 157-164, https://doi.org/10.1007/s10126- 003-0017-5.
Dong, S. R., Kong, J., Zhang, T. S., Meng, X. H., and Wang, R. C., 2006. Parentage determination of Chinese shrimp () based on microsatellite DNA markers., 258: 283-288, https://doi.org/10.1016/j.aquacul ture.2006.04.044.
FAO, 2018.. Rome, 223pp.
Fishback, A. G., Danzmann, R. G., Ferguson, M. M., and Gibson, J. P., 2002. Estimates of genetic parameters and genotype by environment interactions for growth traits of the rainbow trout () as inferred using molecular pedigrees., 206: 137-150, https://doi.org/10.1016/S0044-8486 (01)00707-4.
Fjalestad, K. T., Gjedrem, T., Carr, W. H., and Sweeney, J., 1997. The shrimp breeding program. Selective breeding of. AKVAFORSK, Report no 17/97, 85pp.
Fjalestad, K. T., Moen, T., and Gomez-Raya, L., 2003. Prospects for genetic technology in salmon breeding programmes., 34: 397-406, https://doi.org/10.1046/j.1365- 2109.2003.00823.x.
Fu, J., Shen, Y., Xu, X., and Li, J., 2016. Genetic parameter es- timates for growth of grass carp,, at 10 and 18 months of age., 450: 342-348, https:// doi.org/10.1016/j.aquaculture.2015.08.018.
Garcia, D. K., and Alcivar-Warren, A., 2007. Characterization of 35 new microsatellite genetic markers for the Pacific whiteleg shrimp,: Their usefulness for studying genetic diversity of wild and culture stocks, tracing pedigree in breeding programs and linkage mapping., 26: 1203-1216, https://doi.org/10.2983/0730- 8000(2007)26[1203:CONMGM]2.0.CO;2.
Gheyas, A. A., Woolliams, J. A., Taggart, J. B., Sattar, M. A., Das, T. K., McAndrew, B. J., and Penman, D. J., 2009. Heritability estimation of silver carp () harvest traits using microsatellite based parentage assignment., 294: 187-193, https://doi.org/10.1016/j.aquacul ture.2009.06.013.
Gilmour, A. R., Gogel, B. J., Cullis, B. R., and Thompson, R., 2009. ASReml User Guide Release 3.0. VSN.
Gitterle, T., Johansen, H., Erazo, C., Lozano, C., Cock, J., and Salazar, M., 2007. Response to multi trait selection for harvest body weight, overall survival, and resistance to white spot syndrome virus (WSSV) in()., 272 (Suppl 1): S262, https://doi.org/10.1016/j.aquaculture. 2007.07.073.
Gitterle, T., Rye, M., Salte, R., Cock, J., Johansen, H., Lozano, C., Suarez, J. A., and Gjerde, B., 2005. Genetic (co)variation in har- vest body weight and survival in()under standard commercial conditions., 243: 83-92, https://doi.org/10.1016/j.aquaculture.2004.10.015.
Godin, D. M., Carr, W. H., Hagino, G., Seguro, F., Sweeney, J. N., and Blankenship, L., 1996. Evaluation of a fluorescent elasto- mer internal tag in juvenile and adult shrimp., 139: 243-248, https://doi.org/10.1016/0044- 8486(95)01174-9.
Huang, Y., Yin, Z., Weng, S., He, J., and Li, S., 2012. Selectivebreeding and preliminary commercial performance offor resistance to white spot syndrome virus (WSSV)., 364-365: 111-117, https://doi.org/10.1016/j.aqua culture.2012.08.002.
Jerry, D. R., Preston, N. P., Crocos, P. J., Keys, S., Meadows, J. R., and Li, Y., 2004. Parentage determination of Kuruma shrimp()using microsatellite mark- ers (Bate)., 235: 237-247, https://doi.org/10.1016/ j.aquaculture.2004.01.019.
Jerry, D. R., Stewart, T., Purvis, I. W., and Piper, L. R., 2001. Evaluation of visual implant elastomer and alphanumeric internal tags as a method to identify juveniles of the freshwater crayfish,., 193: 149-154, https:// doi.org/10.1016/S0044-8486(00)00477-4.
Kalinowski, S. T., Taper, M. L., and Marshall, T. C., 2007. Revis- ing how the computer program CERVUS accommodates ge- notyping error increases success in paternity assignment., 16: 1099-1106, https://doi.org/10.1111/j.1365- 294X.2007.03089.x.
Karaket, T., Poompuang, S., Na-Nakorn, U., Kamonrat, W., and Hallerman, E. M., 2011. DNA microsatellite-based evaluation of early growth performance among strains of freshwater prawnde Man., 311: 115- 122, https://doi.org/10.1016/j.aquaculture.2010.11.042.
Kenway, M., Matthew, M., Salmon, M., McPhee, C., Benzie, J., Wilson, K., and Knibb, W., 2006. Heritability and genetic cor- relations of growth and survival in black tiger prawnreared in tanks., 259: 138-145, https:// doi.org/10.1016/j.aquaculture.2006.05.042.
Kocour, M., Mauger, S., Rodina, M., Gela, D., Linhart, O., and Van- deputte, M., 2007. Heritability estimates for processing and quality traits in common carp (L.) using a mole-cular pedigree., 270: 43-50, https://doi.org/10.1016/ j.aquaculture.2007.03.001.
Kong, N., Li, Q., Yu, H., and Kong, L., 2015. Heritability estimates for growth-related traits in the Pacific oyster () using a molecular pedigree., 46: 499-508, https://doi.org/10.1111/are.12205.
Kristjánsson, T., and Arnason, T., 2016. Heritability of econo- mically important traits in the Atlantic codL., 47: 349-356, https://doi.org/10.1111/ are.12496.
Li, D. Y., Kong, J., Meng, X. H., Luan, S., Luo, K., Lu, X., and Cao, B. X., 2016. Development of multiplex PCR systems of microsatellite markers for Pacific white shrimp () and its application for parentage identification.,37: 58-67 (in Chinese with English abstract).
Li, W. J., Luan, S., Luo, K., Sui, J., Xu, X. D., and Tan, J., 2015. Genetic parameters and genotype by environment interaction for cold tolerance, body weight and survival of the Pacific white shrimpat different temperatures., 441: 8-15, https://doi.org/10.1016/j.aquaculture.2015. 02.013.
Liu, P., Kong, J., Shi, T., Zhuang, Z., Deng, J., and Xu, H., 2000. RAPD analysis of wild stock of penaeid shrimp () in the China’s coastal waters of Huanghai and Bohai Seas., 22: 89-93 (in Chinese with English abstract).
Luan, S., Yang, G. L., Wang, J. Y., Luo, K., Zhang, Y. F., and Gao, Q., 2012. Genetic parameters and response to selection for harvest body weight of the giant freshwater prawn.,362-363: 88-96, https:// doi.org/10.1016/j.aquaculture.2012.05.011.
Navarro, A., Zamorano, M. J., Hildebrandt, S., Gines, R., Agui- lera, C., and Afonso, J. M., 2009. Estimates of heritabilities and genetic correlations for growth and carcass traits in gilt- head seabream (L.), under industrial condi- tions.,289: 225-230, https://doi.org/10.1111/are. 12290.
Nguyen, N. H., Quinn, J., Powell, D., Elizur, A., Thoa, N. P., and Nocillado, J., 2014. Heritability for body colour and its genetic association with morphometric traits in Banana shrimp ()., 15: 132, https:// doi.org/10.1186/s12863-014-0132-5.
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., and McAndrew, B. J., 2011. A comparison of communal and separate rearing of families in selective breed- ing of common carp (): Estimation of genetic parameters., 322-323: 39-46, https://doi.org/10. 1016/j.aquaculture.2011.09.031.
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., and McAndrew, B. J., 2013. A comparison of communal and separate rearing of families in selective breed- ing of common carp (): Responses to selec- tion., 408: 152-159, https://doi.org/10.1016/j.aqua culture.2013.06.005.
Nolasco-Alzaga, H. R., Perez-Enriquez, R., Enez, F., Bestin, A., Palacios, E., and Haffray, P., 2017. Quantitative genetic pa- rameters of growth and fatty acid content in the hemolymph of the whiteleg shrimp., 482: 17-23, https://doi.org/10.1016/j.aquaculture.2017.09.015.
Norris, A., Bradley, D. G., and Cunningham, E. P., 2000. Paren- tage and relatedness determination in farmed Atlantic salmon () using microsatellite markers., 182: 73-83, https://doi.org/10.1016/S0044-8486(99)00247-1.
Omasaki, S. K., van Arendonk, J. A. M., Kahi, A. K., and Ko- men, H., 2016. Defining a breeding objective for Nile tilapia that takes into account the diversity of smallholder production systems., 133: 404- 413, https://doi.org/10.1111/jbg.12210.
Pérez-Enriquez, R., and Max-Aguilar, A., 2016. Pedigree trace- ability in whiteleg shrimp () using gene- tic markers: A comparison between microsatellites and SNPs., 42: 227-235, https://doi.org/10.7773/cm.v42 i4.2662.
Robertson, A., 1959. The sampling variance of the genetic corre- lation coefficient., 15: 469-485, https://doi.org/10. 2307/3001774.
Rodzen, J. A., Famula, T. R., and May, B., 2004. Estimation of parentage and relatedness in the polyploid white sturgeon () using a dominant marker approach for duplicated microsatellite loci., 232: 165-182, https:// doi.org/10.1016/S0044-8486(03)00450-2.
Sae-Lim, P., Kause, A., Mulder, H. A., Martin, K. E., Barfoot, A. J., Parsons, J. E., Davidson, J., Rexroad III, C. E., van Arendonk, J. A. M., and Komen, H., 2013. Genotype-by-environ- ment interaction of growth traits in rainbow trout (): A continental scale study., 91: 5572-5581, https://doi.org/10.2527/jas.2012-5949.
Stram, D. O., and Lee, J. W., 1994. Variance components testing in the longitudinal mixed effects model., 50: 1171- 1177, https://doi.org/10.2307/2533455.
Sui, J., Luan, S., Luo, K., Meng, X. H., Lu, X., and Cao, B. X., 2015. Genetic parameters and response to selection for har- vest body weight of Pacific white shrimp,., 1: 1-9, https://doi.org/10.1111/are. 12729.
Tan, J., Luan, S., Luo K., Guan, J. T., Li, W. J., and Sui, J., 2016. Heritability and genotype by environment interactions for growth and survival inat low and high densities., 48: 1430-1438, https://doi. org/10.1111/are.12978.
Valles-Jimenez, R., Cruz, P., and Perez-Enriquez, R., 2004. Popu- lation genetic structure of Pacific white shrimp () from Mexico to Panama: Microsatellite DNA va- riation., 6: 475-484, https://doi.org/10. 1007/s10126-004-3138-6.
Vandeputte, M., Kocour, M., Mauger, S., Dupont-Nivet, M., De Guerry, D., and Rodina, M., 2004. Heritability estimates for growth-related traits using microsatellite parentage assignment in juvenile common carp (L.)., 235: 223-236, https://doi.org/10.1016/j.aquaculture.2003.12.019.
Vandeputte, M., Kocour, M., Mauger, S., Rodina, M., Launay, A., and Gela, D., 2008. Genetic variation for growth at one and two summers of age in the common carp (L.): Heritability estimates and response to selection., 277: 7-13, https://doi.org/10.1016/j.aquaculture.2008.02.009.
Wilson, A. J., Réale, D., Clements, M. N., Morrissey, M. M., Postma, E., and Walling, C. A., 2010. An ecologist’s guide to the animal model.,79: 13-26, https:// doi.org/10.1111/j.1365-2656.2009.01639.x.
. E-mail: luansheng@ysfri.ac.cn
September 2, 2019;
November 4, 2019;
May 14, 2020
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Taxonomy and Phylogeny of a New Marine Planktonic Ciliate, Strombidium pseudorapulum sp. n.(Protozoa, Ciliophora, Oligotrichia)
- Can Langmuir Circulations Solve the Problem of Insufficient Upper-Ocean Mixing?
- Dynamic Diurnal Changes in Green Algae Biomass in the Southern Yellow Sea Based on GOCI Images
- Transcriptomic Profiling of the Immune Response to Crowding Stress in Juvenile Turbot (Scophthalmus maximus)
- A Novel Approach for Evaluating Nonstationary Response of Dynamic Systems to Stochastic Excitation
- Isolation of Enterococcus faecium with Feeding Attractant Function from Pacific White Shrimp (Litopenaeus vannamei) Intestine
