Transcriptomic Profiling of the Immune Response to Crowding Stress in Juvenile Turbot (Scophthalmus maximus)
2020-09-28HUOHuanhuanGAOXiaoqiangFEIFanQINFeiHUANGBinandLIUBaoliang
HUO Huanhuan, GAO Xiaoqiang, FEI Fan, QIN Fei, HUANG Bin,and LIU Baoliang, 2), *
Transcriptomic Profiling of the Immune Response to Crowding Stress in Juvenile Turbot ()
HUO Huanhuan1), 3), GAO Xiaoqiang1), FEI Fan1), QIN Fei1), HUANG Bin1),and LIU Baoliang1), 2), *
1),,,266071,2),,266071,3),,330045,
In this study, juvenile turbotwere vaccinated with attenuated(EIBAV1) and reared at two different densities, low density (LD), (5.25±0.02)kgm−2, as control group and high density (HD), (20.53±0.05)kgm−2, as experimental group. Only density was considered as the variable. Five weeks after vaccination, the transcriptomes of spleen and head kidney from the turbot in two groups were analyzed with RNA-Seq technology. A total of 447 million reads were assembled into 41136 genes with an average length of 1274bp and a N50size of 2295bp. A comparison of gene expression between HD and LD groups revealed 1155 differentially expressed genes (DEGs). Enrichment and pathway analysis of the 10 immune-related DEGs showed the centrality of toll-like receptor signaling pathway, cytosolic DNA-sensing pathway and platelet activation in the host immune responses.The 5 overexpressed inflammatory cytokines and 5 downregulated signal-regulated cytokines genes are covered by these immune-related DEGs. It was inferred that cells suffer damage and the immune response is restrained in turbot under crowding stress.
turbot; transcriptome; crowding stress; immune response
1 Introduction
Crowding stress in teleost may increase the incidence of physical injuries (Juell., 2004; Turnbull., 2005; North., 2006), susceptibility to diseases (Ellis, 2001; Tort, 2011) and suppress to immune response (Nardocci., 2014; Yarahmadi., 2015). Many studies have demonstrated the influence of crowding stress on immune system (Jia., 2016). Lymphocytopenia is a classic hematological response to stress and the number of lymphocytes can be decreased under chronic stressthe suppression of lymphocyte proliferation and the induction of apoptosis by B cells (Pulsford., 1995; Verburg- van., 1999). The stock density has a direct relationship with the decrease in lymphocytes (Cristea., 2012; Zebral., 2015). In addition, crowding stress can inhibit the production of specific antibodies immunoglobulin M (IgM) in fish blood by elevating cortisol level (Var- gas-Chacoff., 2014). The negative effect of crowding stress on IgM production was observed when the sub-Antarctic notothenioid fish () was challenged withprotein extract (Vargas., 2014). Similarly, elevated cortisol level in- hibited IgM production inkept at a crowding stress (Iguchi., 2003). By increasing the pro- duction of relative oxygen species, crowding stress can also affect the level of oxidative stress and alter cellular functions (Neiva., 2010). However, the knowledge of howcrowding stress suppresses the immune response is still limited.
The second-generation sequencing-based whole transcriptome analysis tool, RNA-seq, allows the simultaneous and comprehensive characterization of all gene activities in a specific biological process. Accordingly, RNA- seq has been employed to understand the immune events during a wider perspective on different biological settings in teleost species (Peatman., 2013; Zhu., 2015). RNA-seq allows us not only to identify the gene and char- acterize the gene expression level at the same time but also to elucidate host-bacterial interactions (Chen., 2010). To date, many immune studies in turbot have been performed using RNA-seq following infection (Robledo., 2014; Gao., 2016; Ronza., 2016). Using this sequencing technique, very rich immunity-related genes in several fish species were identified, which included Ja- panese seabass (), turbot (), mud loach () and large yellow croaker () (Xiang., 2010; Patricia., 2012; Long., 2013; Yin., 2016). However, to our knowledge, no research has systematically characterized the immune strategy in turbot following attenuatedvaccination and crowding stress at transcriptomic level.
In farming, the common stocking density of turbot (with an average body weight approximately 150g) is about 10kgm−2. The researches have showed that the ability to withstand density pressure is directly related to the size of fish. The stocking density did not show negative impacts on turbot growth when they are below 14.3kgm−2(body weight approximately 70g), 25.7kgm−2(body weight ap- proximately 180g), 52.3kgm−2(body weight approximately 510g), respectively (Jia., 2016). Edwardsiellosis is a devastating fish disease that causes extensive losses in turbot and the feasibility of using a live attenuated vaccine against edwardsiellosis has been demonstrated to be effective in turbot (Xiao., 2013). In our previous study (Huo., 2017), the special antibody level, serum cortisol level and sIg+cell level in turbot with vaccination are significantly different between (5.25±0.02)kgm−2and (20.53±0.05)kgm−2densities at the 5th week. To gain a better understanding of the effects of crowding stress on the immune response and identify immune-related differentially expressed genes, transcriptomic study was em- ployed in this study. The identification of immune-related genes that are potential markers for disease resistance is a mean of establishing a successful genetic breeding program. An understanding of the immune-related pathways during defense process is a relevant factor of enhancing the resistance of cultured fish to diseases.
2 Materials and Methods
2.1 Fish and Experimental Conditions
Healthy juvenile turbot weighing (150±10)g were obtained from Shandong Oriental Ocean Sci-Tech Co. Ltd. (Shandong, China), where the study was conducted. The fish were acclimated to the experimental environment for 15 days using a flow-through system. The fish were fed twice a day with commercial turbot feed (Ningbo Tech- Bank Co. Ltd., Zhejiang, China) and the daily ration was 1% of their body weight. During the test period, the water temperature was maintained at 16℃±0.5℃ and the salinity at 28.2±3, dissolved oxygen at (8.0±0.5)mgL−2, and pH at 7.5±0.3. The photoperiod was maintained12-h light and 12-h dark.
2.2 Vaccination
The strain (EIBAV1) of attenuatedwas provided by Prof. Yuanxing Zhang and it was described previously by Xiao. (2013). The bacteria were suspended in 0.9% physiological saline at a concentration of approximately 1×107colony forming units (cfu) per mL and stored at 4℃ until use. The experimental fish were injected with 100mL attenuatedand the control received 100mL of 0.9% sterile physiological saline.
After the vaccination with attenuated, all fish were randomly assigned to two different stock densities: LD, 100 fish each tank ((5.25±0.02)kgm−2), and HD, 400 fish each tank ((20.53±0.05)kgm−2). The tank is a cylinder with a diameter of 2m and a height of 1.5m, and the water volume each tank is approximately 3m3. Each density was tested with three replicates. No mortality was observed during the experiment.
2.3 Sampling
In our previous study, 5 fish per tank per week were sampled, and the special antibody IgM in blood in vaccinated turbot approached the maximum at the 5th week (Huo., 2017). Therefore, in this study, 5 fish per tank (totally 15 fish per group) were randomly collected after 5 weeks from each density group immediately after anesthe- tization with 0.05% tricaine methane sulfonate (MS-222, Sigma, St. Louis, MO, USA). The density of HD fish was (26.02±0.05)kgm−2and the density of LD fish was (5.35±0.02)kgm−2at the 5th week. The spleen and head kidney from 15 fish were dissected and pooled (five fish per pool). Samples were flash-frozen in liquid nitrogen and stored at −80℃ prior to RNA extraction.
2.4 RNA Extraction, Library Construction,and Sequencing
Total RNA was extracted from the tissues using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. RNA degradation was monitored in 1% agarose gel. RNA concentration and purity were checked using a Nano-Drop 2000 spectrophotometer (Thermo, NewYork, MA, USA). Furthermore, RNA integrity was assessed with the Agilent 2100 RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). High-quality samples (RNA integrity number>7.0, and RNA concentration>100ngµL−1) were selected for high- throughput sequencing. For each sample, equal amount of RNA from the three replicates were pooled together for RNA-seq library construction.
After total RNA was extracted, mRNA was enriched with oligo (dT) beads. The purified mRNA was fragment- ed into short fragments using fragmentation buffer and then used as a template for first-strand cDNA synthesis using random hexamers and reverse transcriptase. The second- strand cDNA was synthesized using DNA polymerase I and RNase H. Then the cDNA fragments were purified with a QiaQuick PCR extraction kit, end-repaired, added with poly(A), and then ligated to Illumina sequencing adap- ters. The ligation products were size selected with agarose gel electrophoresis and amplified by PCR. Then the cDNA library was constructed from the cDNA synthesized using NEBNext UltraTMRNA Library Prep Kit for Illumina (New England Biolabs [NEB],Ipswich, MA, USA). The cDNA libraries were sequenced using Illumina HiSeqTM4000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
2.5 de novo Assembly of Sequencing Reads
To obtain high-quality clean reads for the transcript as- sembly, raw reads were filtered by removing those contain- ing more than 10% unknown nucleotides; removing low- quality reads containing more than 50% of low-quality bases; removing reads containing adapters. The resulting high-quality sequences wereassembled into contigs and transcripts with Trinity software (http://trinityr- naseq.sf.net) (Grabherr., 2011). To reduce data redundancy, transcripts with a minimum length of 200bp were assembled and mapped to the reference transcriptome using the Bowtie2 short reads alignment tool under default parameters (Li., 2009).
2.6 Gene Annotation
All assembled unigenes were used as queries in sear- ching the NCBI non-redundant (Nr) protein database and SwissProt protein database using the BLASTX program. The cutoff-value was set at 1e−5and only the top gene ID and name were initially assigned to each unigene. The uni- genes were functionally annotated by gene ontology (GO) analysis with Blast2GO software (-value<10−5). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway ana- lysis was performed using KEGG Automatic Annotation Server (KASS) with default parameters (Zhang., 2015).
2.7 Identification of Differentially Expressed Genes
All clean sequencing reads from each of the cDNA libraries were mapped back to the transcriptome assembly using the software Bowtie2 with default settings. The reads aligned to each unigene in the alignment file were counted for each sample. These read counts were normalized as RPKM (reads per kilobase of transcripts per million frag- ments mapped) values (Mortazavi., 2008) and further analysis to identify differentially expressed genes (DEGs) between the two different groups was conducted using a web tool DESeq. The false discovery rate (FDR) method was introduced to determine the threshold-value in mul- tiple tests to judge the significance of the difference in gene expression. If the FDR was <0.05 and there was at least a 2-fold difference in RPKM value between samples, the unigene was considered a significant DEG (Li., 2015).
2.8 Experimental Validation Using qPCR
Twelve DEGs (Table 1) were selected randomly for va- lidation of RNA-seq data by qPCR using a SYBR Premix Ex Taq Kit (Invitrogen) according to the manufacturer’s instructions. The same RNA samples were used for both Illumina library synthesis and the qPCR verification assay. The first strand cDNA was obtained from 2μg of total RNA using a PrimeScript 1st strand cDNA synthesis kit (Takara, Dalian, China). Melting curve analyses were perform- ed following amplification. The specific primers used for qPCR are listed in Table 1, andgene transcript abundance was used as the endogenous control. The thermal profile for SYBR Green qPCR was 95℃ for 30s, fol- lowed by 40 cycles of 95℃ for 5s, 60℃ for 20s, then sub- jected to dissociation curve analysis, 95℃ for 15s, 60℃ for 15s, then up to 95℃ at a rate of 0.1℃s−1, to verify the specificity of the amplicons. The relative gene expression was analyzed using the comparative threshold cycle me- thod established by Livak. (2001).

Table 1 Primers used for qPCR validation of RNA-Seq
3 Results
3.1 de novo Assembly and Annotation
To obtain the turbot transcriptome expression profile after vaccination, four cDNA libraries were constructed using spleen and head kidney from the two groups of HD and LD turbot. The results of transcriptome have been submitted to NCBI (SRA accession no. SRP129900). A total of 447 million clean reads were obtained for subsequent analysis after eliminating low-quality sequences and adaptor sequences from the original data sequence by quality analysis. The clean reads showed a high quality with a Q20 of 97.81% and Q30 of 94.83% (Table 2). Transcriptassembly was performed for the clean reads by Trinity. The total number and length of unigenes were 41136and 52423775bp, respectively. The maximal length of the unigenes was 23178bp with an average length of 1274bp (N50: 2295), and the GC content was 49.13% (Table 3). These results indicated that the sequencing data were high- quality, and the unigenes could be used for subsequent annotation analysis.
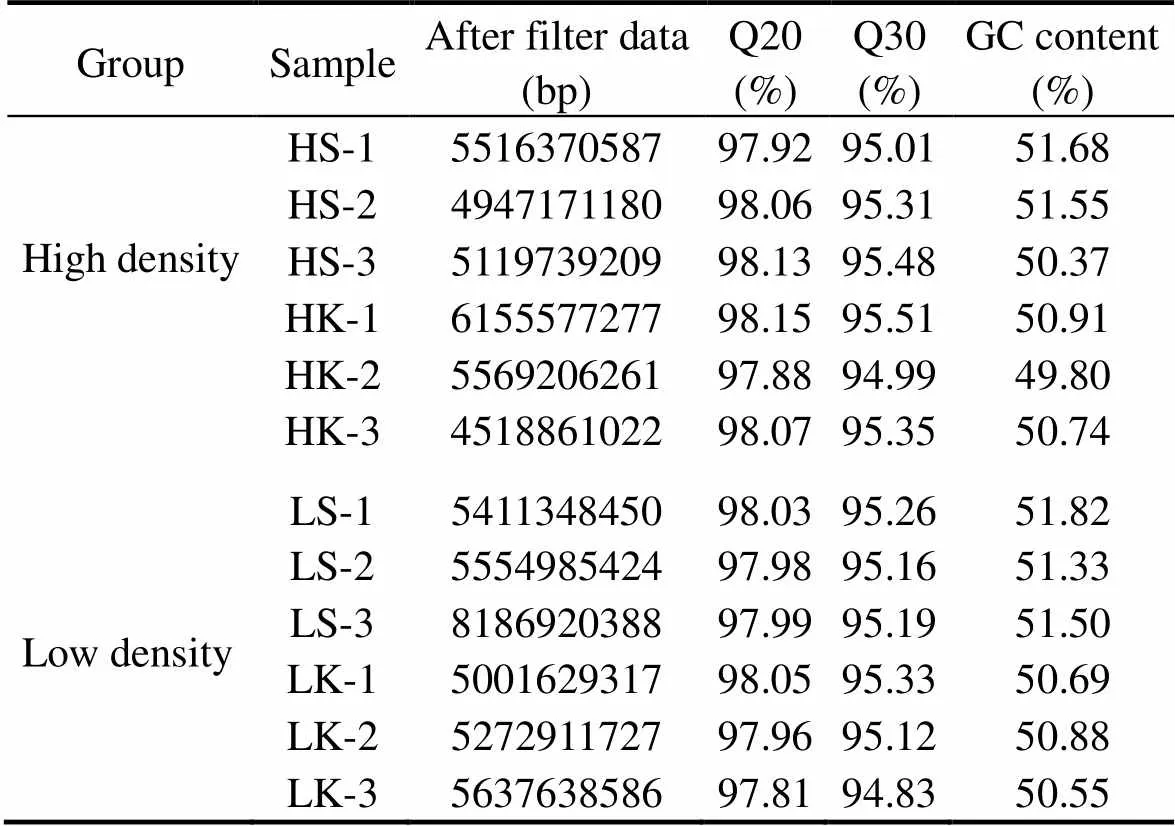
Table 2 The quality of data
Notes: HS and HK represent the spleen and head kidney from high density respectively; LS and LK represent the spleen and head kidney from low density respectively.

Table 3 Summary of de novo assembly of transcriptomic profiles of S. maximus
To obtain comprehensive gene information, 41136 uni- genes were annotated by four databases including Nr, Clus- ters of orthologous groups for eukaryotic complete geno- mes (KOG), SwissProt and KEGG (Table 4). Annotation was matched successfully for 22931 of the unigenes (22806 in Nr, 20101 in SwissProt, 16035 in KOG, and 14332 in KEGG) and 18205 unigenes were without annotation.The distribution of 22931 annotated unigenes from KEGG, KOG, Nr and SwissProt data are shown in Fig.1. In Nr annotation, 22806 unigenes were matched to multiple species ge- nomes, including(31.58%),(25.76%),(5.68%),(5.16%),(4.92%), and others (26.88%) (Fig.2).

Table 4 The statistics of annotated unigenes of transcriptomic profiles in four databases
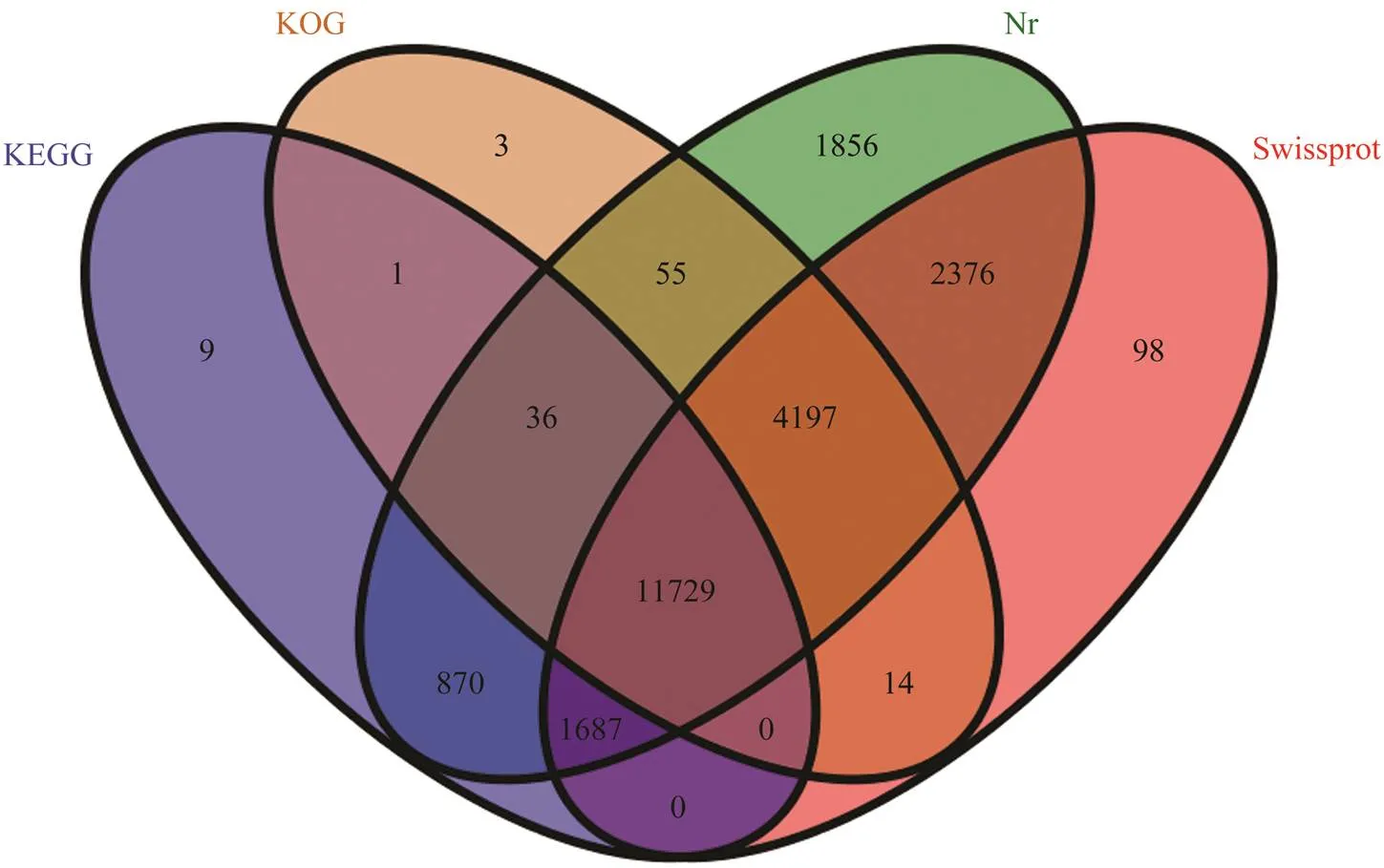
Fig.1 The venn of four database annotation. The distribution of a total 22931 annotated unigenes in KEGG, KOG, Nr and Swissprot database. The blue oval represents KEGG database. The yellow oval represents KOG database. The green oval represents Nr database. The red oval represents Swissprot database. The digital represents the number of annotated unigenes by the corresponding database. The digital at the intersection represents annotated by several corresponding databases at the same time.

Fig.2 The top 10 species match to unigenes of S. maximus. The Y-axis indicates the number of unigenes and the X-axis indicates matched species. From left to right, the matching degree from high to low.
3.2 Identification and Analysis of DEGs
All unigenes were analyzed with DEG-Seq, whose thres- hold was restricted to-value<0.005, |log2 (fold-change)|>1. A total of 1155 genes showed statistically significant differential expressions in two immunologic organs that were compared in HD and LD turbots. In detail, 397 unigenes were identified as DEGs containing 146 significantly upregulated and 251 significantly downregulated unigenes in spleen. Additionally, 758 unigenes were identified asDEGs containing 100 significantly upregulated and 658 sig- nificantly downregulated unigenes in head kidney (Fig.3). In the statistical figures, we observed that the downregu- lation levels of DEGs were significantly higher than the upregulation levels after attenuatedadministration. Among the 1155 DEGs, a group of 84 DEGs was shared between the two organs, 16 up- and 66 down-regu- lated, and 2 either up- or down-regulated depending on organ.

Fig.3 The statistics of different expression gene in spleen (HS vs LS) and head kidney (HK vs LK). The red and the green represent upregulated and downregulated expressed genes, respectively. The digital represents the number of different expression genes. There were 146 upregulated ex- pressed genes and 251 downregulated expressed genes in spleen. There were 100 upregulated expressed genes and 658 downregulated expressed genes in head kidey.
3.3 GO Enrichment and Pathway Analysis
To further investigate the function of the DEGs, they were classified into three GO categories including biological process, molecular function and cellular component, by Blast2GO. A total of 36 GO terms including 16 biological process terms, 12 cellular component terms and 8 molecular functions terms were associated with spleen DEGs (Fig.4A). A total of 42 GO terms including 19 bio- logical process terms, 13 cellular component terms, and 10 molecular functions terms were associated with head kidney DEGs (Fig.4B). Analysis of level 2 GO term distribution showed that cellular process, binding and cell were the most common annotation terms in three GO ca- tegories. The immune system process was retained for fur- ther pathway analysis although it was not the most significant one.
A total of 401 unigenes were homologous to immune- relevant genes based on KEGG annotation and were classi- fied into 15 functional pathways (Table 5). Furthermore, through immune-related DEG screening, 10 DEGs (Table 6) were assigned to six immune-relevant pathways inclu- ding the RIG-I-like receptor signaling pathway, cytoso- lic DNA-sensing pathway, toll-like receptor (TLR) signa- ling pathway,NOD-like receptor signaling pathway, che- mokine signaling pathway, and platelet activation. Among these immune-relevant pathways, we focused on signify- cantly changed signaling pathways (<0.05) such as the TLR signaling pathway, cytosolic DNA-sensing pathway and platelet activation.

Fig.4 Gene ontology (GO) enrichment analysis of the differently expressed genes. According to GO, each differential gene was assigned to corresponding term.The red represents upregulated expressed genes and the green represents downregulated expressed genes. A: ‘HS vs LS’ represents spleen group; A total of 36 GO terms including 16 biological process terms, 12 cellular component terms and 8 molecular functions terms in spleen. B: ‘HK vs LK’ represents head kidney group; A total of 42 GO terms including 19 biological process terms, 13 cellular component terms and 10 molecular functions terms in head kidney.

Table 5 KEGG pathway enrichment analyses of the immune related differently expressed genes and annotation genes

(continued)
Notes: The symbol ‘–’ represents the number of differently expressed genes were zero. The number of annotation genes represents expressed genes in the corresponding pathway in this study.

Table 6 The information of immune related DEGs
Notes: Bold values indicate significantly changed relative to the low density groups. The positive numbers indicate up-regulation and the negative numbers indicate down-regulation.
3.4 Validation of RNA-seq Profiles by qPCR
To validate the RNA-seq data, 12 DEGs were selected randomly for qPCR. Although the tested unigenes display- ed different expression levels (Fig.5), in general, the qPCR results showed a positive correlation with RNA-seq data, indicating the reliability and accuracy of the RNA-seq ex- pression analysis.

Fig.5 Validation of RNA-seq data by using qPCR. X-axis, pairwise comparison groups; Y-axis, fold change in gene expression. CCL20, CISH, IFNA, SOCS3, IL1R2 andPIK3Rwere detected in head kidney; CXCL10, EPOR, CCL19,CXCL13, IL18 and IL1β were detected in spleen. The qPCR data were given in mean±SD, n=3.
4 Discussion
RNA-seq technology uses RNA-seq sequence data to pro- vide a complete picture of the transcriptome under study (global annotation) and simultaneously provide quantitative data related to differential gene expressions. In the present study, we have obtained the transcriptome comparative data ofspleen and head kidney under two different culture densities and have acquired 41136 unigenes after assembling and filtration. Although the tur- bot genome was released and was available in the European Nucleotide Archive, the mapping percentage was very low; therefore, we used the BLASTX program to annotate the unigenes in an approach adopted by other studies (Gao., 2016).
In Nr annotation, only 1123 unigenes were matched to(4.92%) genomes. The origin of flatfish has been the focus of long-term controversy (Ro- bledo., 2016).Phylogenetic studies show low interfamily/suborder statistical support among flatfish species, especially between the Psettoidei and Pleuronectoidei, and the low phylogenetic support is considered the evidence of a polyphyletic origin (Campbell., 2014). Genetic evi- dence on the diversification of flatfish has been reported using genome sequencing of the turbot and the tongue sole (Chen., 2014; Antonio., 2016), suggesting that different strategies are adapted for demersal life (Ro- bledo., 2016).
A total of 1155 DEGs were categorized into many ca- tegories based on GO annotation and pathway analyses. These categories included the immune system, which is the key category we focus on. Below we highlight three key constituents of the immune system, the TLR signaling pathway, the cytosolic DNA-sensing pathway, and platelet activation, which were all influenced significantly by crowding stress.
4.1 TLR Signaling Pathway
Recognition of pathogen-associated molecular patterns (PAMPs) is essential for the activation of innate immunity and it is carried out by toll-like receptors ()., specific families of pattern recognition receptors, are responsible for detecting microbial pathogens and generating in- nate immune responses. Pathogen recognition byprovokes rapid activation of innate immunity byinducing the production of pro-inflammatory cytokines and upregulation of costimulatory molecules as well as pri- ming the adaptive immune system (Pasare., 2004; Werling., 2009; Liu., 2010). Efficient immune responses depend on a close interaction between the innate and adaptive immune systems.serve as an im- portant link between the innate and adaptive immune responses (de Souza., 2008). Importantly,parti- cipate in the direct regulation of the adaptive immune response, possibly as costimulatory molecules (Liu., 2010). Dendritic cells stimulated directly or indirectly by TLRs from pathogens mature into a specific form. They are able to activate a specific immune response that is ap- propriate for the elimination of the pathogen (Dulay., 2009). In the process of recovery from diseases, TLRsalso participate in inflammation and immune responsesthat are driven by self-, allo- or xeno-antigens (Liu., 2010).
In our study, inflammatory cytokines such as interleu- kin (IL)-1β and interferon (IFN)-awere markedly upre- gulated in spleen when turbot suffered crowding stress. It is well-known that the protective properties of these inflammatory factors remove the adverse stimulus in the first line of an organism’s defense against pathogen invasion, but a stronger inflammatory response can lead to tissue damage as well (Du., 2017). Chemokines have a broad spectrum of effects on innate immunity. It can be a promoter to adhere various types of leukocyte to inflammatory loci in early immune response (Bagasra., 1997; Yoshie., 2001). Research has shown that turbotchemokines were dramatically and rapidly induced at 5h after challenge and the expression ofchemokine was observed only at 12h after the challenge (Liu., 2007). In our study, the overexpression ofchemokine ligand-9 () andchemokine ligand-10 () were observed at the 5th week in high density groups, which may indicate the spleen was damaged. In addition, we found that phosphoinositide 3- kinase receptors () was markedly downregulated in spleen. Phosphoinositide 3-kinase () inhibition leads to the suppression ofbut inducedexpression (Marshall., 2012). The PI3ks are lipid kinases that are activated by receptor tyrosine. They can suppress cell apoptosis, promote cell growth, and play important roles in the immune response (Shi., 2010).Downregulatedsignifies that crowding stress is disadvantageous to the immune response.
In head kidney transcriptomic data, no inflammatory cytokines and costimulatory molecules were observed to be upregulated or downregulated. However,and mitogen-activated protein kinase1 () were drama- tically downregulated in the TLR signaling pathway. The downregulation ofindicates the suppression of extracellular signal-regulated kinases (), which are widely expressed protein kinase intracellular signaling mole- cules involved in maintaining cell survival and mediating intracellular signal transduction in response to a variety of stimuli (Ikeyama., 2002; Dou., 2005; Rehani., 2009). Some studies show that upregulatedplays a key role in environmental adaptation processes, which is helpful for protecting cells from stresses (Helmreich., 2001; Padmini., 2014). In our study, downregulatedsignifies that crowding stress damages spleen cells.
4.2 Cytosolic DNA-Sensing Pathway
Specific families of pattern recognition receptors are re- sponsible for detecting foreign DNA from invading microbes or host cells, and generating innate immune responses.Cytosolic DNA sensors detect intracytoplasmic double-stranded DNA and signal through a central adaptor protein, stimulator ofgenes (STING), which recruits and activates the downstream TBK1/IRF3 signaling axis to induce the production of type I(Wu., 2014; Roers., 2016). Type Iare critical for innate immune defense against viral infections (Schneider., 2014; Yang., 2015). Furthermore, cytosolic DNA also activates STING-dependent and STING-inde- pendent autophagy to promote host defense against bacterial infection (Liang., 2014; Watson., 2015). Studies by Rengaraj. (2016) have shown that the cytosolic DNA-sensing pathway is the most important im- mune pathway in necrotic enteritis-induced chickens. Si- milar results were obtained in our study. The DEG enrichment in the cytosolic DNA-sensing pathway are significantly upregulated in spleen.,, andare upregulated, butis downregulated in the cytosolic DNA-sensing pathway. Previous studies have confirmed thatis important for regulating the Th1 and Th2 immune responses (Nakanishi., 2001). The down-regulation ofwill attenuate the immune response ability of the spleen cell. In head kidney, DEGs are not enriched in the cytosolic DNA-sensing pathway, indicating that spleen and head kidney play different roles in the immune response.
4.3 Platelet Activation
Platelets are not only cell fragments that plug the leak in a damaged blood vessel, but also key components in the innate immune system, which is supported by the pre- sence ofon platelets (Cox., 2011). Recently platelets have been reported to express,and, reinforcing their role as primitive immune cells in host defense (Cognasse., 2005; Aslam., 2006). As platelets are usually the first cells to respond to a wound, they play an important role in the immune response to infection through their activation (Fitzgerald., 2006; Yeaman., 2010). Platelets act as components of the innate immune system, and can detect the presence of in- fectious agents and coordinate the response to the patho- gen (Garraud., 2010). When activated, platelets se- crete over 300 proteins (Coppinger., 2004), including the bioactive molecules adenosine diphosphate () and serotonin to reduce blood loss, cytokines and chemo- kine recruiting leucocytes to deal with any potential infection, and antimicrobial peptides to kill pathogens.The activation of platelets leads to the secretion of antimicrobial peptides, although many bacteria have become resistant to these peptides (Yeaman., 1998).
In our study,and talin () were significant- ly downregulated in the platelet activation pathway in head kidney.is a master regulator of platelet inte- grin activation. If there is a lack of, the plate- let aggregation function will be damaged severely and in- tegrin αIIbβ3 activation will also be inhibited (Griffiths., 2001; Han., 2006).downregulation decreases ligand binding to cellular integrins, resulting in re- strained platelet and leukocyte function., the se- cond messenger cyclic adenosine monophosphate (cAMP) and cAMP-dependent protein kinase A are important mediators in determining the response of a cell to external stimuli (Grandoch., 2010; Bhattacharjee., 2012). Activatedincreases the intracellular cAMP level, which enhances the host defense (Fritz., 2004).The downregulation ofwill attenuate the response of a cell to external stimuli by decreasing cAMP levels.
In addition to the nine genes mentioned above, gene DEAD-box 3 X () is significantly downregulated in the RIG-I-like receptor signaling pathway.par- ticipates in regulating mRNA translation and some signaling pathways (Cruciat., 2013; Fullam., 2013).directly regulates Wnt/β-catenin signaling and INF- induced signal pathways (Thompson., 2011; Cruciat., 2013).is responsible for detecting viral pa- thogens and generating innate immune responses. Vacci- nia virus protein K7 can combine with DDX3X to suppress the innate immune response (Oda, 2009). Downregulatedindicates that the innate immune response is inhibited.
5 Conclusions
Using RNA-seq-based transcriptome profiling, we assessed, for the first time, the transcriptomic responses of turbot cultured in different densities following vaccination with attenuated. A total of 1155 DEGs were annotated against four databases. We identified immune- related DEGs through comparing the gene expression le- vels in high- and low-stock densities. After RNA-seq data analysis and qPCR validation, we observed that immune- related DEGs were primarily enriched in the TLR signaling pathway and cytosolic DNA-sensing pathway in spleen while platelet activation occurred in head kidney. The in- flammatory cytokines,,, and, and signal-regulated cytokines,,,, and,are involved in these pathways. Overexpressed inflammatory cytokines and downregulated signal-regu- lated cytokines show that cells suffer damage and the im- mune response is restrained when turbot is stressed by crowding. Although further studies are needed to characterize the key immune factors governing the turbot immune responses, these results can provide a foundation to evaluate the relationship between immunosuppression and crowding stress.
Acknowledgements
This work was supported by the National Key R&D Pro- gram of China (No. 2017YFB0404001), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (No. 2017HYZD04), the Qingdao Shinan District Science and Technology Bureau (No. 2016-3-006) and the Modern Agriculture Industry System Construction Special Funds (No. CARS-47-G24).
Antonio, F., Diego, R., André, C., Miguel, H., Patricia, P., Juan, R., Jèssica, G. G., Laia, C., Xabier, B., Marta, G., Marina, M. H., Gabriel, F. C., Beatriz, G., José, L. G., José, L. A. F., Belen, G. P., Xoana, T., Carlos, F., Anna, V., Antonio, H. P., Roderic, G., José, A. Á. D., Antonio, G. T., Ana, V., Xulio, M., Toni, G., Beatriz, N., Carmen, B., Tyler, A., and Paullino, M., 2016. Whole genome sequencing of turbot (; Pleuronectiformes): A fish adapted to demersal life., 23 (3): 181-192.
Aslam, R., Speck, E. R., Kim, M., Andrew, R. C., Bang, K. W. A., Nestel, F. P., Ni, H., Lazarus, A. H., Freedman, J., and Sem- ple, J. W., 2006. Platelet Toll-like receptor expression modulates lipopolysaccharide-induced thrombocytopenia and tumor necrosis factor-α production., 107 (2): 637-641.
Bagasra, O., 1997. The immunobiology of interferon-gamma in- ducible protein 10kD (IP-10): A novel, pleiotropic member of the C-X-C chemokine superfamily., 8 (3): 207-219.
Bhattacharjee, R., Xiang, W., Wang, Y., Zhang, X. Y., and Billiar, T. R., 2012. cAMP prevents TNF-induced apoptosis through inhibiting DISC complex formation in rat hepatocytes., 423 (1): 85-90.
Campbell, M. A., Chen, W. J., and López, J. A., 2014. Molecular data do not provide unambiguous support for the monophyly of flatfishes (Pleuronectiformes): A reply to Betancur-R and Ortí., 75 (1): 149-153.
Chen, S. L., Liu, Y., Dong, X. L., and Meng, L., 2010. Cloning, characterization, and expression analysis of a CC chemokine gene from turbot ()., 36 (2): 147-155.
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., Song, W., An, N., Chalopin, D., Volff, J. N., Hong, Y., Li, Q., Sha, Z., Zhou, H., Xie, M., Yu, Q., Liu, Y., Xiang, H., Wang, N., Wu, K., Yang, C., Zhou, Q., Liao, X., Yang, L., Hu, Q., Zhang, J., Zhu, Y., Du, M., Zhao, Y., Schartl, M., Tang, Q., and Wang, J., 2014. Whole-genome sequence of a flatfish pro- vides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle., 46 (3): 253.
Cognasse, F., Hamzeh, H., Chavarin, P., Acquart, S., Genin, C., and Garraud, O., 2005. Evidence of Toll-like receptor molecules on human platelets., 83 (2): 196-198.
Coppinger, J. A., Cagney, G., Toomey, S., Kislinger, T., Belton, O., Mcredmond, J. P., Cahill, D. J., Emili, A., Fitzgerald, D. J., and Maguire, P. B., 2004. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions., 103 (6): 2096-2104.
Cox, D., Kerrigan, S. W., and Watson, S. P., 2011. Platelets and the innate immune system: Mechanisms of bacterial-induced platelet activation., 9 (6): 1097.
Cristea, V., Mocanu, M., Antache, A., Docan, A., Dediu, L., Ion, Sandita., and Coadă, M. T., 2012. Effect of stocking density on leuckocyte reaction of(Walbaum, 1792)., 45 (2): 31- 36.
Cruciat, C. M., Dolde, C., de Groot, R. E., Ohkawara, B., Rein- hard, C., Korswaqen, H. C., and Niehrs, C., 2013. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in wnt- β-catenin signaling., 339 (6126): 1436-1441.
de Souza, A. L., and Seguro, A. C., 2008. Two centuries of me- ningococcal infection: From vieusseux to the cellular and mole- cular basis of disease., 57 (11): 1313-1321.
Dou, F., Yuan, L. D., and Zhu, J. J., 2005. Heat shock protein 90 indirectly regulates ERK activity by affecting RAF protein me- tabolism., 37 (7): 501- 505.
Du, X., Li, Y., Li, D., Lian, F., Yang, S., Wu, J., Liu, H., Bu, G., Meng, F., Cao, X., Zeng, X., Zhang, H., and Chen, Z., 2017. Transcriptome profiling of spleen provides insights into the antiviral mechanism in, after poly (i:c) challenge., 62: 13-23.
Dulay, A. T., Buhimschi, C. S., Zhao, G., Oliver, E. A., Mbele, A., Jing, S. C., and Buhimschi, I. A., 2009. Soluble TLR2 is present in human amniotic fluid and modulates the intraam- niotic inflammatory response to infection., 182 (11): 7244-7253.
Ellis, A. E., 2001. Innate host defense mechanisms of fish a- gainst viruses and bacteria., 25 (8): 827-839.
Fitzgerald, J. R., Foster, T. J., and Cox, D., 2006. The interaction of bacterial pathogens with platelets., 4 (6): 445-457.
Fritz, J. H., Brunner, S., Birnstiel, M. L., Buschle, M., Gabain, A., Mattner, F., and Zauner, W., 2004. The artificial antimi- crobial peptide KLKLLLLLKLK induces predominantly a TH2- type immune response to co-injected antigens., 22 (25- 26): 3274-3284.
Fullam, A., and Schröder, M., 2013. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication., 1829 (8): 854-865.
Gao, C., Qiang, F., Su, B., Zhou, S., Liu, F., Song, L., Zhang, M., Ren, Y., Dong, X., Tan, F., and Li, C., 2016. Transcriptomic profiling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot () follow- ing, challenge., 65: 159-168.
Garraud, O., and Cognasse, F., 2010. Platelet toll-like receptor expression: The link between ‘danger’ ligands and inflammation., 9 (5): 322-333.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., Adiconis, X., Fan, L., Raychowdhury, R., Zeng, Q., Chen, Z., Mauceli, E., Hacohen, N., Gnirke, A., Rhind, N., Palma, F. D., Birren, B. W., Nusbaum, C., Toh, K. L., Friedman, N., and Regev, A., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome., 29 (7): 644-652.
Grandoch, M., Roscioni, S. S., and Schmidt, M., 2010. The role of epac proteins, novel camp mediators, in the regulation of immune, lung and neuronal function., 159 (2): 265-284.
Griffiths, E. K., Krawczyk, C., Kong, Y. Y., Raab, M., Hyduk, S. J., Bouchard, D., Chan, V. S., Kozieradzki, I., Oliveira-Dos- Santos, A. J., Wakeham, A., Ohashi, P. S., Cybulsky, M. I., Rudd, C. E., and Penninger, J. M., 2001. Positive regulation of T cell activation and integrin adhesion by the adapter Fyb/Slap., 293 (5538): 2260-2263.
Han, J., Lim, C. J., Watanabe, N., Soriani, A., Ratnikow, B., Cal- derwood, D. A., Puzon-Mclaughlin, W., Lafuente, E. M., Bou- ssiotis, V. A., Shattil, S. J., and Ginsberg, M. H., 2006. Recon- structing and deconstructing agonist-induced activation of integrin αIIbβ3., 16 (18): 1796-1806.
Helmreich, E. J. M., 2001.Oxford University Press, New York, 368pp.
Huo, H., Yin, S., Jia, R., Huang, B., Lei, J., and Liu, B., 2017. Effect of crowding stress on the immune response in turbot () vaccinated with attenuated., 67 (5): 353-358.
Iguchi, K., Ogawa, K., Nagae, M., and Ito, F., 2003. The influence of rearing density on stress response and disease susceptibility of ayu ()., 220 (1): 515-523.
Ikeyama, S., Kokkonen, G., Shack, S., Wang, X. T., and Holbrook, N. J., 2002. Loss in oxidative stress tolerance with aging linked to reduced extracellular signal-regulated kinase and Akt kinase activities., 16 (1): 114-116.
Jia, R., Liu, B. L., Feng, W. R., Han, C., Huang, B., and Lei, J. L., 2016. Stress and immune responses in skin of turbot () under different stocking densities., 55: 131-139.
Juell, J. E., and Fosseidengen, J. E., 2004. Use of artificial light to control swimming depth and fish density of Atlantic sal- mon () in production cages., 233 (1- 4): 269-282.
Li, G., Zhao, Y., Liu, Z., Gao, C., Yan, F., Liu, B., and Feng, J., 2015.assembly and characterization of the spleen tran- scriptome of common carp () using illumina paired-end sequencing., 44 (2): 420-429.
Li, R., Yu, C., Li, Y., Lam, T., Yiu, S., Kristiansen, K., and Wang, J., 2009. Soap2: An improved ultrafast tool for short read a- lignment., 25 (15): 1966-1967.
Liang, Q., Seo, G. J., Choi, Y. J., Kwak, M., Ge, J., Rodgers, M. A., Shi, M., Leslie, B. J., Hopfner, K., Ha, T., Oh, B., and Jung, J. U., 2014. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses., 15 (2): 228-238.
Liu, G., Zhang, L., and Zhao, Y., 2010. Modulation of immune responses through direct activation of toll-like receptors to T cells., 160 (2): 168-175.
Liu, Y., Chen, S. L., Meng, L., and Zhang, Y. X., 2007. Cloning, characterization and expression analysis of a novel CXC che- mokine from turbot ()., 23 (4): 711-720.
Livak, K. J., and Schmittgen, T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod., 25 (4): 402-408.
Long, Y., Li, Q., Zhou, B., Song, G., Li, T., and Cui, Z., 2013.assembly of mud loach () skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion., 8 (2): e56 998.
Marshall, N. A., Galvin, K. C., Corcoran, A. M., Boon, L., Hiqqs, R., and Mills, K. H., 2012. Immunotherapy with PI3K inhibitor and toll-like receptor agonist induces IFN-γ+IL-17+polyfunctional T cells that mediate rejection of murine tumors., 72 (3): 581-591.
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq., 5 (7): 621-628.
Nakanishi, K., Yoshimoto, T., Tsutsui, H., and Okamura, H., 2001. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu., 12 (1): 53-72.
Nardocci, G., Navarro, C., Cortés, P. P., Imarai, M., Montoya, M., Valenzuela, B., Jara, P., Castillo, C. A., and Fernánde, R., 2014. Neuroendocrine mechanisms for immune system regulation during stress in fish., 40 (2): 531-538.
Neiva, B., Ronaldolima, D. L., Bernardo, B., Alcirluiz, D., and Alexpires, D. O. N., 2010. Growth, biochemical and physiological responses ofwith different stock- ing densities and handling., 301 (1-4): 22-30.
North, B. P., Turnbull, J. F., Ellis, T., Port, M. J., Migaud, H., Bron, J., and Bromage, N. R., 2006. The impact of stocking density on the welfare of rainbow trout ()., 255 (1-4): 466-479.
Oda, S., Schröder, M., and Khan, A. R., 2009. Structural basis for targeting of human RNA helicase DDX3 by poxvirus protein K7., 17 (11): 1528-1537.
Padmini, E., and Tharani, J., 2014. Fourier transform infrared spec- troscopic study on HSP70 and ERK in fish hepatocytes during pollution induced stress., 4 (2): 114-125.
Pasare, C., and Medzhitov, R., 2004. Toll-like receptors: Linking innate and adaptive immunity., 6 (15): 1382- 1387.
Patricia, P., Pablo, B., Alejandro, R., Sonia, D., Gabriel, F., Berta, F., Josep, V. P., Sergi, B., Beatriz, N., and Antonio, F., 2012. High-throughput sequence analysis of turbot () transcriptome using 454-pyrosequencing for thediscovery of antiviral immune genes., 7 (5): e35369.
Peatman, E., Li, C., Peterson, B. C., Straus, D. L., Farmer, B. D., and Beck, B. H., 2013. Basal polarization of the mucosal com- partment insusceptible and resistant channel catfish ()., 56 (4): 317-327.
Pulsford, A. L., Crampe, M., Langston, A., and Glynn, P. J., 1995. Modulatory effects of disease, stress, copper, TBT and vitamin E on the immune system of flatfish., 5 (8): 631-643.
Rehani, K., Wang, H., Garcia, C. A., Kinane, D. F., and Martin, M., 2009. Toll-like receptor-mediated production of IL-1Ra is negatively regulated by GSK3the MAPK ERK1/2., 182 (1): 547-553.
Rengaraj, D., Truong, A. D., Lee, S. H., Lillehoj, H. S., and Hong, Y. H., 2016. Expression analysis of cytosolic DNA-sensing pathway genes in the intestinal mucosal layer of necrotic enteritis-induced chicken., 170: 1-12.
Robledo, D., Hermida, M., Rubiolo, J. A., Fernández, C., Blanco, A., Bouza, C., and Martínez, P., 2016. Integrating genomic re- sources of flatfish (Pleuronectiformes) to boost aquaculture production., 21: 41-55.
Robledo, D., Ronza, P., Harrison, P. W., Losada, A. P., Bermúdez, R., Pardo, B. G., Redondo, M. J., Bobadilla, A. S., Quiroga, M. I., and Martínez, P., 2014. RNA-Seq analysis reveals significant transcriptome changes in turbot () suffering severe enteromyxosis., 15 (1): 1149.
Roers, A., Hiller, B., and Hornung, V., 2016. Recognition of en- dogenous nucleic acids by the innate immune system., 44 (4): 739-754.
Ronza, P., Robledo, D., Bermúdez, R., Losada, A. P., Pardo, B. G., Bobadilla, A. S., Quiroga, M. I., and Martínez, P., 2016. RNA- Seq analysis of early enteromyxosis in turbot (): New insights into parasite invasion and immune evasion strategies., 46 (8): 507-517.
Schneider, W. M., Chevillotte, M. D., and Rice, C. M., 2014. Interferon-stimulated genes: A complex web of host defenses., 32 (1): 513-545.
Shi, Z., Hodges, V. M., Dunlop, E. A., Percy, M. J., Maxwell, A. P., EI-Tanani, M., and Lappin, T. R., 2010. Erythropoietin- induced activation of the JAK2/STAT5, PI3K/Akt, and Ras/ ERK pathways promotes malignant cell behavior in a modified breast cancer cell line., 8 (4): 615-626.
Thompson, M. R., Kaminski, J. J., Kurtjones, E. A., and Fitzgerald, K. A., 2011. Pattern recognition receptors and the innate immune response to viral infection., 3 (6): 920- 940.
Tort, L., 2011. Stress and immune modulation in fish., 35 (12): 1366-1375.
Turnbull, J., Bell, A., Adams, C., Bron, J., and Huntingford, F., 2005. Stocking density and welfare of cage farmed atlantic salmon: Application of a multivariate analysis., 243(1-4): 121-132.
Vargas-Chacoff, L., Martínez, D., Oyarzún, R., Nualart, D., Ola- varría, V., Yáñez, A., Bertrán, C., Ruiz-Jarabo, I., and Mancera, J. M., 2014. Combined effects of high stocking density andtreatment on the immune system, metabolism and osmoregulatory responses of the sub-antarc- tic notothenioid fish., 40 (2): 424-434.
Verburg-van Kemenade, B. L., Nowak, B., Engelsma, M. Y., and Weyts, F. A. A., 1999. Differential effects of cortisol on apop- tosis and proliferation of carp B-lymphocytes from head kidney, spleen and blood., 9 (5): 405-415.
Watson, R., Bell, S., Macduff, D., Kimmey, J., Diner, E., Olivas, J., Vance, R., Stallings, C., Virgin, H., and Cox, J., 2015. The cytosolic sensor cGAS detects, DNA to induce type I interferons and activate autophagy., 17 (6): 811.
Werling, D., Jann, O. C., Offord, V., Glass, E. J., and Coffey, T. J., 2009. Variation matters: TLR structure and species-speci- fic pathogen recognition., 30 (3): 124- 130.
Wu, J., and Chen, Z. J., 2014. Innate immune sensing and signaling of cytosolic nucleic acids., 32 (1): 461.
Xiang, L. X., He, D., Dong, W. R., Zhang, Y. W., and Shao, J. Z., 2010. Deep sequencing-based transcriptome profiling analysis of bacteria-challengedreveals insight into the immune-relevant genes in marine fish., 11 (472): 1-21.
Xiao, J., Chen, T., Liu, B., Yang, W., Wang, Q., Qu, J., and Zhang, Y., 2013.mutant disrupted in type III se- cretion system and chorismic acid synthesis and cured of a plasmid as a live attenuated vaccine in turbot., 35 (3): 632-641.
Yang, K., Wang, J., and Wu, M., 2015. Mesenchymal stem cells detect and defend against gammaherpesvirus infectionthe cGAS-STING pathway., 5 (7820): 1-9.
Yarahmadi, P., Miandare, H. K., Hoseinifar, S. H., Gheysvandi, N., and Akbarzadeh, A., 2015. The effects of stocking density on hemato-immunological and serum biochemical parameters of rainbow trout ()., 23 (1): 55-63.
Yeaman, M. R., 2010. Bacterial-platelet interactions: Virulence meets host defense., 5 (3): 471-506.
Yeaman, M. R., Bayer, A. S., Koo, S. P., Foss, W., and Sullam, P. M., 1998. Platelet microbicidal proteins and neutrophil defen- sin disrupt thecytoplasmic membrane by distinct mechanisms of action., 101 (1): 178-187.
Yin, F., Gao, Q., Tang, B., Sun, P., Han, K., and Huang, W., 2016. Transcriptome and analysis on the complement and coagulation cascades pathway of large yellow croaker () to ciliate ectoparasiteinfection., 50: 127-141.
Yoshie, O., Imai, T., and Nomiyama, H., 2001. Chemokines in im- munity., 78 (78): 57-110.
Zebral, Y. D., Zafalon-Silva, B., Mascarenhas, M. W., and Robaldo, R. B., 2015. Leucocyte profile and growth rates as indicators of crowding stress in pejerrey fingerlings ()., 46 (9): 2270-2276.
Zhang, X., Wang, S., Chen, S., Chen, Y., Liu, Y., Shao, C., Wang, Q., Lu, Y., Gong, G., Ding, S., and Sha, Z., 2015. Transcriptome analysis revealed changes of multiple genes involved in immunity induringinfection., 43 (1): 209-218.
Zhu, J., Li, C., Ao, Q., Tan, Y., Luo, Y., Guo, Y., Lan, G., Jiang, H., and Gan, X., 2015. Trancriptomic profiling revealed the signatures of acute immune response in tilapia (), following, challenge., 46 (2): 346-353.
. E-mail: Liubl@ysfri.ac.cn
May 20, 2019;
September 17, 2019;
April 16, 2020
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Dynamic Diurnal Changes in Green Algae Biomass in the Southern Yellow Sea Based on GOCI Images
- Nutrient Enrichment Regulates the Growth and Physiological Responses of Saccharina japonica to Ocean Acidification
- Isolation of Enterococcus faecium with Feeding Attractant Function from Pacific White Shrimp (Litopenaeus vannamei) Intestine
- Taxonomy and Phylogeny of a New Marine Planktonic Ciliate, Strombidium pseudorapulum sp. n.(Protozoa, Ciliophora, Oligotrichia)
- Reducing the Common Environmental Effect on Litopenaeus vannamei Body Weight by Rearing Communally at Early Developmental Stages and Using a Reconstructed Pedigree
- Can Langmuir Circulations Solve the Problem of Insufficient Upper-Ocean Mixing?
