Cloning and Bioinformatics Analysis of pepck Gene in Vibrio alginolyticus
2020-09-28FuyuanZENGYinZHAOShihuiZHOUChuanhaoPANMiaoXIEHuanyingPANG
Fuyuan ZENG, Yin ZHAO, Shihui ZHOU, Chuanhao PAN, Miao XIE, Huanying PANG*
1. Shenzhen Institute of Guangdong Ocean University, Shenzhen 510000, China; 2. College of Fisheries, Guangdong Ocean University/Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals/Key Laboratory of Disease Control for Aquatic Economic Animals, Department of Education of Guangdong Province, Zhanjiang 524088, China
Abstract [Objectives] To clone the pepck gene of Vibrio alginolyticus strain HY9901 and analyze its sequence by bioinformatics. [Methods] According to the complete gene sequence of V. alginolyticus on GenBank, specific primers were designed to amplify the target gene pepck by PCR. The sequence of the pepck gene was analyzed using bioinformatics. The phylogenic tree of pepck gene and the corresponding single-subunit three-dimensional structure were constructed. [Results] The pepck gene of V. alginolyticus strain HY9901 has a full length of 1 629 bp, with theoretical molecular weight of 60.12 kD. The prediction results show that there is no signal peptide or transmembrane region at the N-terminus of the sequence, the amino acid sequence contains 11 phosphorylation sites of casein kinase II. The prediction results of protein subcellular localization indicate that PEPEK protein is localized in the cytoplasm. The protein is stable and hydrophobic. The tertiary structure of the PEPCK protein of V. alginolyticus is similar to that of Vibrio parahaemolyticus. It is predicted that PEPCK has a major functional domain PEPCK_ATP. In the secondary structure, alpha helix, random coil, and extended strand accounted for 21.96%, 52.03% and 26.01%, respectively. The PEPCK homology between V. alginolyticus and Vibrio diabolicus is as high as 99%. [Conclusions] This study lays the foundation for further understanding the function of pepck gene in V. alginolyticus.
Key words Vibrio alginolyticus, pepck gene, Gene cloning, Bioinformatics analysis
1 Introduction
Phosphoenolpyruvate carboxykinase (PEPCK) is a key metabolic enzyme in the process of gluconeogenesis, and it catalyzes oxaloacetate (OAA) into phosphoenolpyruvate (PEP) and carbon dioxide (CO2) and is necessary for gluconeogenesis in almost all organisms[1-2]. According to OAA’s inorganic phosphate donor, PEPCK can be divided into three types: ATP-dependent, GTP-dependent and PPi (inorganic pyrophosphate, PPi)-dependent[3-5]. The metabolic processes that the protein participates in have a very important role in the blood glucose level[6], virulence regulation[7], antiviral immunity[8]and other aspects of the organism. PEPCK has different physiological roles in the carbon metabolism of different organisms[9]. InMycobacteriumtuberculosis, the PEPCK protein is encoded by thepckAgene, and it can stimulate the body to produce an immune response and is a good vaccine candidate molecule[10-11].
Vibrioalginolyticus(Vibrionaceae:Vibrio) is a kind of halophilic and mesophilic gram-negative bacillus. It is widely distributed in the ocean, estuaries, mariculture ponds and other water environments[12], and is the main pathogen of fish, shrimp and shellfish, bringing huge economic losses to the marine aquaculture industry[13-15]. At the same time, the bacterium is a zoonotic pathogen that can contaminate human food, causing diarrhea, trauma infection, otitis media and other diseases[16-18]. Antibiotics are the main chemical drugs used to prevent and treatV.alginolyticusinfections. However, the problems of drug residues and bacterial resistance caused by long-term use of antibiotics are becoming more and more serious, so there is an urgent need to develop new vaccines to prevent and treatV.alginolyticusdiseases[19]. Studies have shown that the PEPCK protein ofV.alginolyticusis closely related to drug resistance[20], and it is identified as the cross immunogenic protein ofV.alginolyticus,VibrioharveyiandVibrioparahaemolyticus[21]. To further study the function ofpepckgene ofV.alginolyticus, with the help of bioinformatics software, a phylogenetic tree for the PEPCK protein ofV.alginolyticuswas constructed, and its basic physical and chemical properties, signal peptide and transmembrane structure, secondary structure and tertiary structure were predicted and analyzed in this paper, in order to lay a foundation for the development of the PEPCK subunit vaccine ofV.alginolyticus.
2 Materials and methods
2.1 Strains and vectorThe virulent strainV.alginolyticusHY9901 was preserved by the Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals[22].Escherichiacolistrain DH5α was preserved by Shenzhen Institute of Guangdong Ocean University. The cloning vector pMD18-T was purchased from Takara Biotechnology Co., Ltd.
2.2 Main reagentsThe reagents used included RxTaq DNA polymerase, DNA Marker (Takara, Japan), bacterial genomic DNA extraction kit, DNA gel recovery kit (TransGen, China). Other reagents of analytical grade were imported or produced in China. The mass concentration of the antibiotic ampicillin (Ap) was 100 μg/mL.
2.3 Apparatus and equipmentPCR instrument (Bio-Rad, Canada; TaKaRa, Japan), electrophoresis apparatus (Liuyi, China), high-speed refrigerated centrifuge (Eppendorf, America), constant-temperature incubator (Boxun, China) and clean bench (Huilong, China).
2.4 Extraction of total DNA ofVibrioalginolyticusstrain HY9901V.alginolyticuswas cultured according to the method of Pang Huanying[23]. The genomic DNA ofV.alginolyticuswas extracted according to the instructions of the bacterial genomic DNA extraction kit and store at -20 ℃ for later use.
2.5 Cloning ofpepckgeneBased on the complete gene sequence ofV.alginolyticuson Genbank, a pair of primers was designed: 5′-ATGACCGTTATGGAACAT-3′ (forward primer P1) and 5′-ATCAATCTGAGGACCAGC-3′ (reverse primer P2). PCR reaction program was as follows: pre-denaturation at 95 ℃ for 5 min; denaturation at 95 ℃ for 30 s, anneal at 62 ℃ for 30 s; extension at 72 ℃ for 90 s, 33 cycles; and extension at 72 ℃ for 10 min. A small amount of the PCR product was sampled and examined by 1% agarose gel electrophoresis.
2.6 Purification of PCR productsThe PCR product was purified according to the instructions of the DNA gel recovery kit.
2.7 Ligation and transformationThe PCR product was ligated to the pMD18-T vector, placed in a refrigerator at 4 ℃ overnight, and transformed intoE.coliDH5α competent cells. The main steps of transformation were as follows: (i) Adding 5 μL of ligation product to 50 μL of DH5α competent cells; (ii) Ice bath for 30 min, water bath (42 ℃) for 90 s, then placing on ice for 3-5 min; (iii) Adding 1 000 μL of LB medium and shaking culture at 37 ℃, 200 r/min for 1 h; (iv) Centrifuging at 3 000 r/min for 2 min at room temperature, discarding the supernatant and retaining 100 μL, suspending the bacterium gently and spreading on LB plate (containing 100 μg/mL of Ap); (v) Letting stand for 30 min and inverting in an incubator (37 ℃) for 10-12 h.
2.8 Picking and sequencing of positive clonePositive monoclones on the plate were picked, placed in 1 mL LB medium (containing 100 μg/mL of Ap), shaking-cultured at 200 r/min for 4-5 h at 37 ℃, and sent to Shanghai Bioengineering Service Co., Ltd. for sequencing.
2.9 Bioinformatics analysis of PEPCKNCBI and DNAMAN were used for sequence homology comparison and similarity analysis. Using ExPASy Proteomics Server program, the physical and chemical properties of PEPCK were analyzed. The amino acid sequence of the PEPCK protein ofV.alginolyticuswas analyzed to determine molecular weight (Mw) and theoretical isoelectric point (pI); ORF Finder software was used to predict the open reading frame (ORF) ofpepcksequence; SignalP 4.0 Server was used to predict signal peptide sequence; TMHMM Server 2.0 was used to predict the transmembrane domain; SoftBerry-Psite software was used to predict the functional site distribution of amino acid sequence; PSORT II Prediction was used to predict subcellular location; Clastal 2.0 and MEGA 5.0 software were used to construct NJ phylogenetic tree; and SWISS-MODEL program was used to build three-dimensional structure.
3 Results
3.1 Cloning ofpepckgeneAfter PCR amplification, a specific band of 1 629 bp was obtained (Fig.1A). After PCR amplification of positive clone, a band of 1 629 bp was also obtained (Fig.1B). The sequencing analysis shows that thepepckgene has an open reading frame (ORF) of 1 629 bp, encoding 542 amino acids (Fig.2). The sequence of the gene was submitted to GenBank, with registration number MT683849.

Note: A: M, DL2000 DNA molecular marker; 1-4, PCR product. B: M, DL2000 DNA molecular marker; 1-2, PCR product of positive clones.
3.2 Physical and chemical properties of PEPCK proteinThe total atom number of the PEPCK protein ofV.alginolyticusstrain HY9901 is 8 390, and its molecular structure is C2682H4156N708O825S19, with theoretical molecular weight of 60.127 89 kDa and theoretical pI of 5.03. The instability coefficient is 26.78, less than the threshold value of 40, indicating that the protein is stable. The fat coefficient is 76.83. The total average coefficient of hydrophilicity is -0.363. The protein does not contain selenocysteine (Sec) or pyrrolysine (Pyl). The total number of acidic amino acids (Asp+Glu) is 80, and the total number of basic amino acids (Arg+Lys) is 57. The N-terminal is methionine (Met). The half-life of expression in yeast andE.coliis greater than 20 and 10 h, respectively, and inin-vitromammalian reticulocytes, the half-life is 30 h.
3.3 Sequence analysis of PEPCK proteinSignalP 4.0 Server was used to predict the structure of N-terminal signal peptide, and no signal peptide was found. The prediction result of TMHMM Server 2.0 shows that there was no transmembrane domain. The prediction result of SoftBerry-Psite shows that the amino acid sequence contains four N-glycosylation sites, one glycosaminoglycan attachment site, one cAMP- and cGMP-dependent protein kinase phosphorylation site, 11 protein kinase C phosphorylation sites, 11 casein kinase II phosphorylation sites, two tyrosine kinase phosphorylation sites, 10 N-myristoylation sites, one amidation site, 12 microbodies C-terminal targeting signal sites, one cell attachment sequence, one ATP/GTP-binding site motif A and one phosphoenolpyruvate carboxykinase (ATP) signal site (Fig.2). The prediction results of protein subcellular localization indicate that PEPCK protein is located in the cytoplasm.
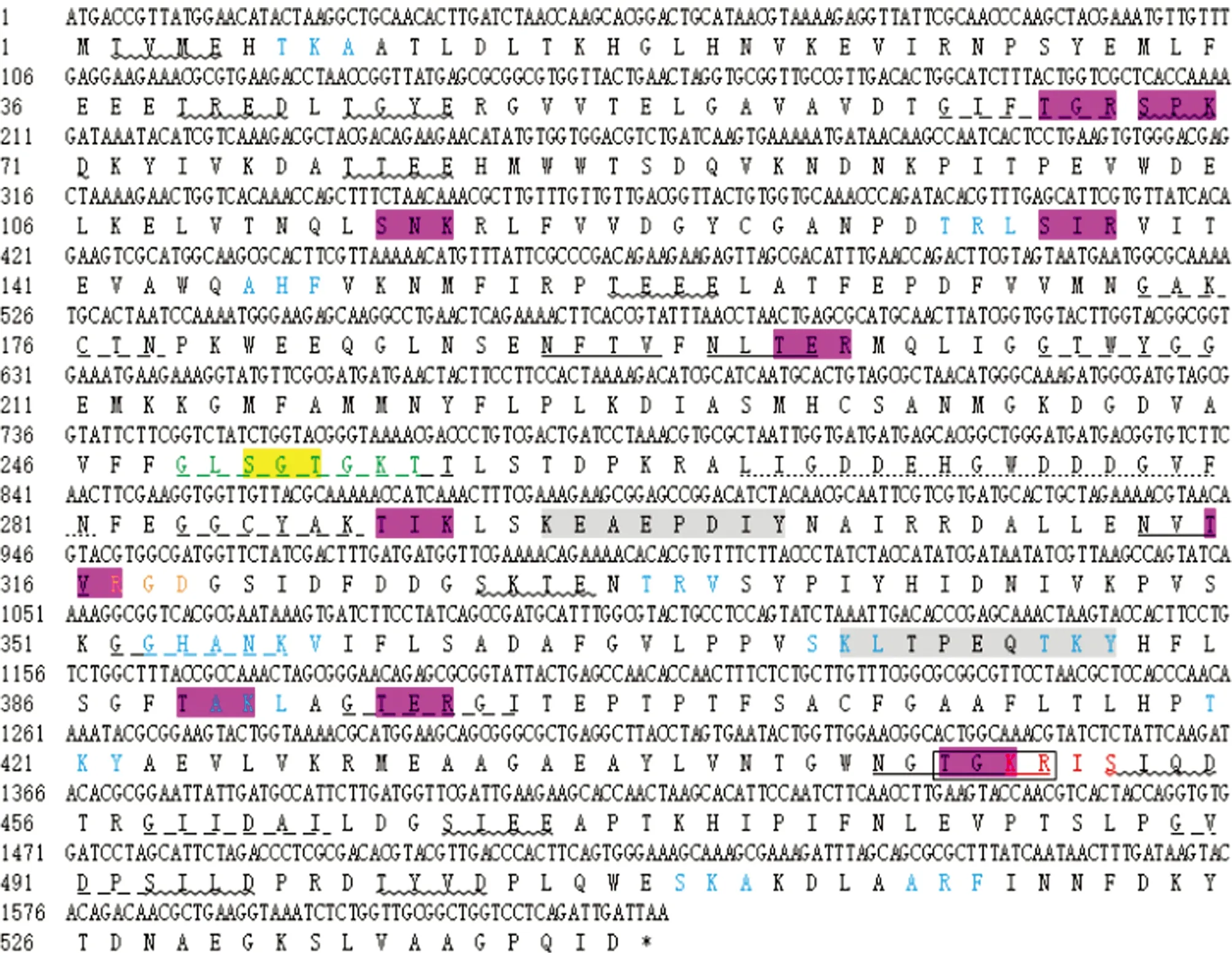
Note: "*" represents a terminator; "_" marks N-glycosylation site; "" marks casein kinase II phosphorylation site; "_ _" marks N-myristoylation site; "▭" marks amidation site; "" marks phosphoenolpyruvate carboxykinase (ATP) signal site; red font marks cAMP- and cGMP-dependent protein kinase phosphorylation site; blue font marks microbodies C-terminal targeting signal site; orange font marks cell attachment sequence; green font marks ATP/GTP-binding site motif A; yellow background marks glycosaminoglycan attachment site; pink background marks protein kinase C phosphorylation site; and gray background marks tyrosine kinase phosphorylation site.
3.4 Analysis of the homology and evolution of PEPCKBLAST analysis found that PEPCK ofV.alginolyticushas high homology with those of other bacteria. Among them, the homology with the PEPCK amino acid sequence of V. diabolicus was 99.08%. The comparison result of DNAMAN software shows that the PEPCK in Vibrio is highly conserved (Fig.3). The phylogenetic tree shows that the PEPCK proteins ofV.alginolyticusstrain HY9901 and Vibrio sp.11-4(1) are clustered together (Fig.4).
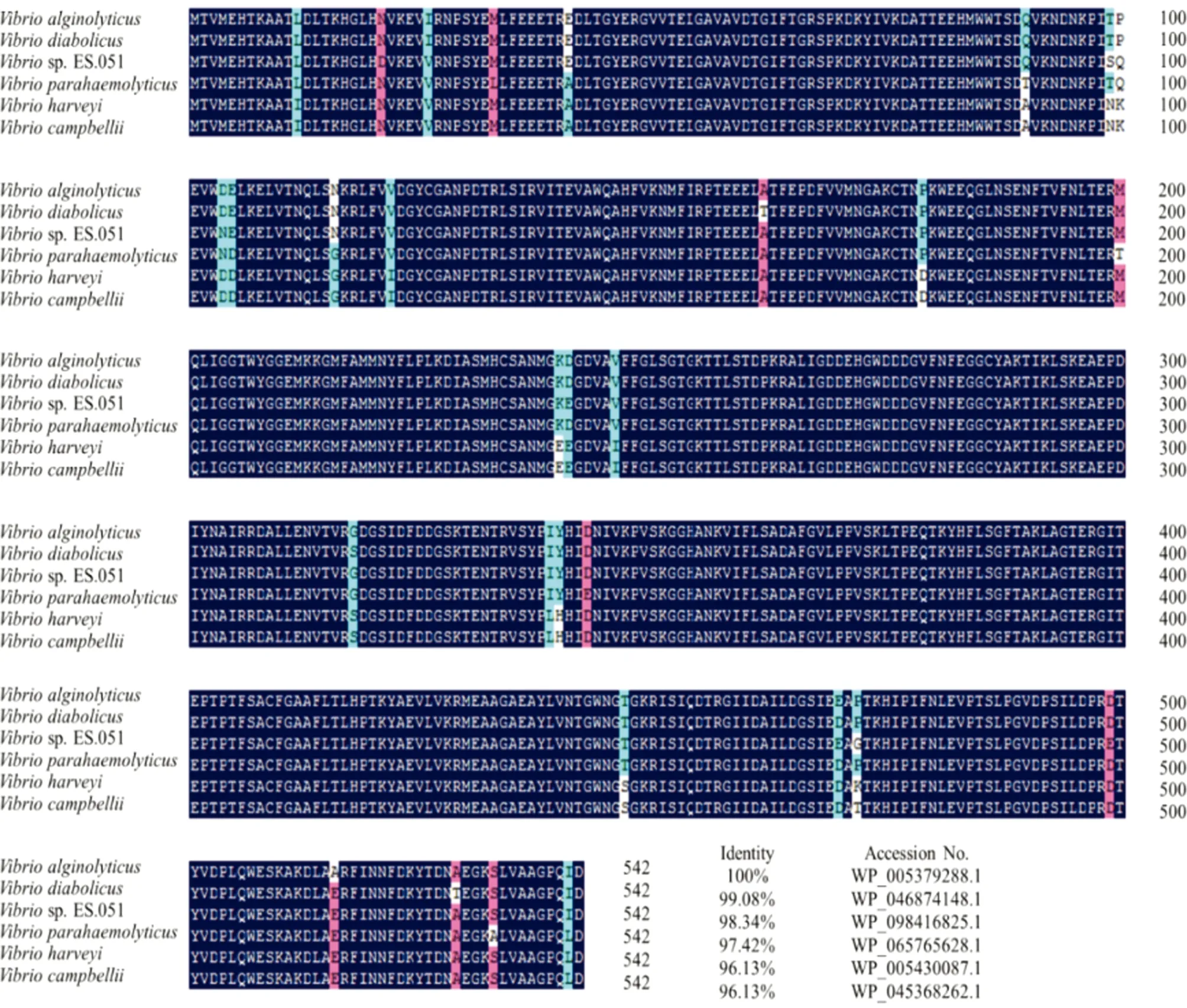
Fig.3 Similarity comparison between PEPCK amino acid sequences
3.5 Prediction of function domain and secondary structure of PEPCKThe functional domain of PEPCK protein was predicted using SMART, and it is found to have a functional domain of PEPCK_ATP (23-493aa) (Fig.5). The secondary structure of PEPCK protein was predicted using SOPMA. The result shows that alpha helix accounts for 21.96%, random coil accounts for 52.03%, and extended strand accounts for 26.01%.

Fig.5 Domain prediction of PEPCK protein

Note: blue, alpha helix; purple, random coil; red, extended strand.
3.6 Subunit structure of PEPCKThe amino acid sequence of PEPCK was submitted to the SWISS-MODEL program, and many similar homologous sequences were obtained through database search. Among them, one or several were chosen as templates. After comparison and adjustment, the single-subunit tertiary structure model of PEPCK was constructed (Fig.7A). After comparison, it was found that PEPCK subunit ofV.alginolyticushas a similar configuration with that ofV.parahaemolyticus(Fig.7B). (V.alginolyticus: QMEAN, -0.47; Cβ, -0.48; All atom, -0.25; solvation, 0.80; torsion, -0.63;V.parahaemolyticus: QMEAN, -0.26; Cβ, -0.37; All atom, -0.24; solvation, 0.71; torsion, -0.41).

Note: A: V. alginolyticus strain HY9901; B: V. parahaemolyticus.
4 Discussion
This study successfully cloned thepepckgene of phosphoenolpyruvate carboxykinase inV.alginolyticusstrain HY9901. The gene is 1 629 bp in length. The amino acid sequence of PEPCK encoded has the highest homology with that inV.diabolicus, up to 99%. The homology with otherVibriospecies is higher than 96%, indicating that PEPCK has high conservation. This is consistent with its role as a common immunogenic protein ofV.alginolyticus,V.harveyiandV.parahaemolyticus[21]. It is known that PEPCK ofM.tuberculosishas good immunogenicity and antigenicity, and it may be used as one of a group of antigens for serological diagnosis of tuberculosis[24]. In the future, PEPCK ofV.alginolyticuscan be verified through prokaryotic expression, immunogenicity and immune protection tests to develop new highly effective subunit vaccines for the prevention and treatment of vibriosis. Phylogenetic tree analysis is consistent with amino acid sequence homology alignment. The PEPCK proteins ofV.alginolyticusstrain HY9901 andVibriosp.11-4(1) are grouped together. The molecular weight of the PEPCK protein ofV.alginolyticusis 60.127 89 kDa, and it is a non-hydrophilic stable protein. Studies have shown that ATP-dependent PEPCK can exist in bacteria[3], so it is reasonable that the prediction result shows that the PEPCK protein ofV.alginolyticushas a PEPCK_ATP functional domain. The secondary structure contains three conformations of α-helix, random coil and extended strand. The prediction results of SWISS-MODEL show that the tertiary structure of the single subunit ofV.alginolyticusPEPCK has a similar configuration to that of the single subunit ofV.parahaemolyticusPEPCK. It is speculated that the two have similar functions.
At present, there are few studies on the function ofpepckgene ofV.alginolyticus. In this study, the bioinformatics of PEPCK protein is analyzed, and its physical and chemical properties and structural characteristics are successfully predicted, providing a foundation for subsequent verification and functional research. In the future, thepepckgene inV.alginolyticuscan be knocked out to further explore the mechanism of action ofV.alginolyticuspepckgene in drug resistance and immunity.
杂志排行
Asian Agricultural Research的其它文章
- Impact of COVID-19 Epidemic on the International Food Supply Chain and Countermeasures of Shandong Province
- Study of Factors Influencing China-ASEAN Agricultural Product Trade Development in the Context of "the Belt and Road"
- Comprehensive Evaluation on the Level of Agricultural Economic Development in Hubei Province Based on Principal Component Analysis
- Effect of Foliar Application of Selenium Fertilizer on Yield, Selenium Content and Heavy Metal Contents of Waxy Maize
- Effects of Applying Trace Clements on Yield and Quality of Flue-cured Tobacco Cuibi-1
- Analysis of the Correlation between Middle-season Rice Yield and Meteorological Factors in Jingdong County from 2009 to 2016
