The Morphology and Phylogeny of Three Diophrys Ciliates Collected from the Subtropical Waters of China, Including a New Species (Ciliophora; Euplotia)
2020-09-27ZHANGChaojianHUANGJieYETingtingLUBorongandCHENXiangrui
ZHANG Chaojian, HUANG Jie, YE Tingting, LU Borong, and CHEN Xiangrui, *
The Morphology and Phylogeny of ThreeCiliates Collected from the Subtropical Waters of China, Including a New Species (Ciliophora; Euplotia)
ZHANG Chaojian1), #, HUANG Jie2), #, YE Tingting1), LU Borong3), 4), and CHEN Xiangrui1), *
1),,315211,2),,,430072,3),,266003,4),,266003,
Threespecies,n. sp.,Borror, 1965 and(Dujardin, 1841) Kahl, 1932, were identified from the subtropical brackish waters in China, their morphology was investigated using live observation and protargol impregnation.n. sp. can be distinguished from the congeners by four ellipsoidal macronuclear nodules. Phylogenetic analyses show that the three species with single marginal cirrus are closely related, namelyn. sp.,Song., 2009 andHu, 2008. Our population ofcorresponds very well with previous populations in both living morphology and ciliature. However, the SSU rDNA sequences of three well-described populations differ from each other by 11 to 18 nucleotides, indicating that there might be cryptic species in. Moreover, we provided a detailed description of the symbiotic bacteria/archaeobacteria (?) concentrated on the back surface ofbased on the present population, which was either overlooked or misinterpreted as cortical granules previously.
; morphology; new species; phylogeny; taxonomy
1 Introduction
The-complex is a common group of order Eu- plotida and belongs to the most morphologically complex class Spirotrichea. These species have dominant oral field and strong cirri on ventral side, and generally exist in marine and estuarine biotopes (Song., 2007, 2009a;Lynn, 2008; Hu., 2019). In the past 40 years, this com- plex group was clarified many times and include six genera now. For example, Jankowski (1979) divided it intoand; Hill and Borror (1992) established the third genus,; Jankowski (2007) reassigned these three genera in Diophryinae. Hereafter, three more genera,,and, have been reported and added to this subfa- mily (Jiang and Song, 2010; Jiang., 2011). Subsequently, Huang. (2012) and Fan. (2013) reveal- ed some opinions based on molecular and morphogenetic characters, and some of them did not completely support the classification of the subfamily,.,andwere not included in this group.
After several revisions and divestitures, several species have been removed from the genusThe remain- ing species are mainly characterized by the combined fea- tures, namely three caudal cirri located in a prominent pos- terior concave area, oral area with prominent adoral zone of membranelles and distinct paroral and endoral membranes, ventral side arranged with five frontal, two ventral, five transverse, and one or two left marginal cirri. So far, only ten species still remain in genus:Ruinen, 1938;Borror, 1965;Bor- ror, 1965;Song., 2009a;(Du-jardin, 1841) Kahl, 1932;(Ehrenberg, 1838) Schewiakoff, 1893;Shen., 2011;Hu, 2008;Hu., 2012;Luo., 2014 (Dujardin, 1841; Kahl, 1932; Curds and Wu, 1983; Hill and Borror, 1992; Song and Packroff, 1997; Song and Wilbert, 1994, 2002; Song., 2007, 2009a, b; Hu, 2008;Shen., 2011; Hu., 2012; Fan., 2013; Luo., 2014). Till now, seven out of ten species have been investigated based on both mor- phological descriptions and molecular data.
As a new contribution, the present work describes threeciliates collected from subtropical brackish waters, namelyn. sp.,and. Moreover, the phylogeny of all-com- plex species based on small-subunit rDNA (SSU rDNA) sequences and a key to all valid species ofaccording to morphological characters are provided.
2 Material and Methods
2.1 Sample Collection, Observation and Terminology (Figs.1A–D1)
n. sp. (Fig.1B1) was collected on August 23, 2014 from Songlanshan beach, Xiangshan Island, Ningbo (29˚26΄22΄΄E, 121˚58΄24΄΄N), China (Figs.1A,B). The particle size of sand was 0.15–0.25mm. The upper 10–20cm layer of sand was collected with seawater from the site. The water temperature was about 30℃ and the salinity was 20.
(Fig.1C1) was collected on 15 November 2018 from a brackish water lake (Figs.1A, C, C’) which is connected with the Xiangshan Bay, Ningbo (29˚45΄28΄΄E, 121˚54΄53΄΄N), China. The water tempera- ture was about 19℃ and the salinity was about 22. Samples were taken from the surface layer of lake-bed sediment using a Pasteur pipette and then diluted with untreated water from the collection site.
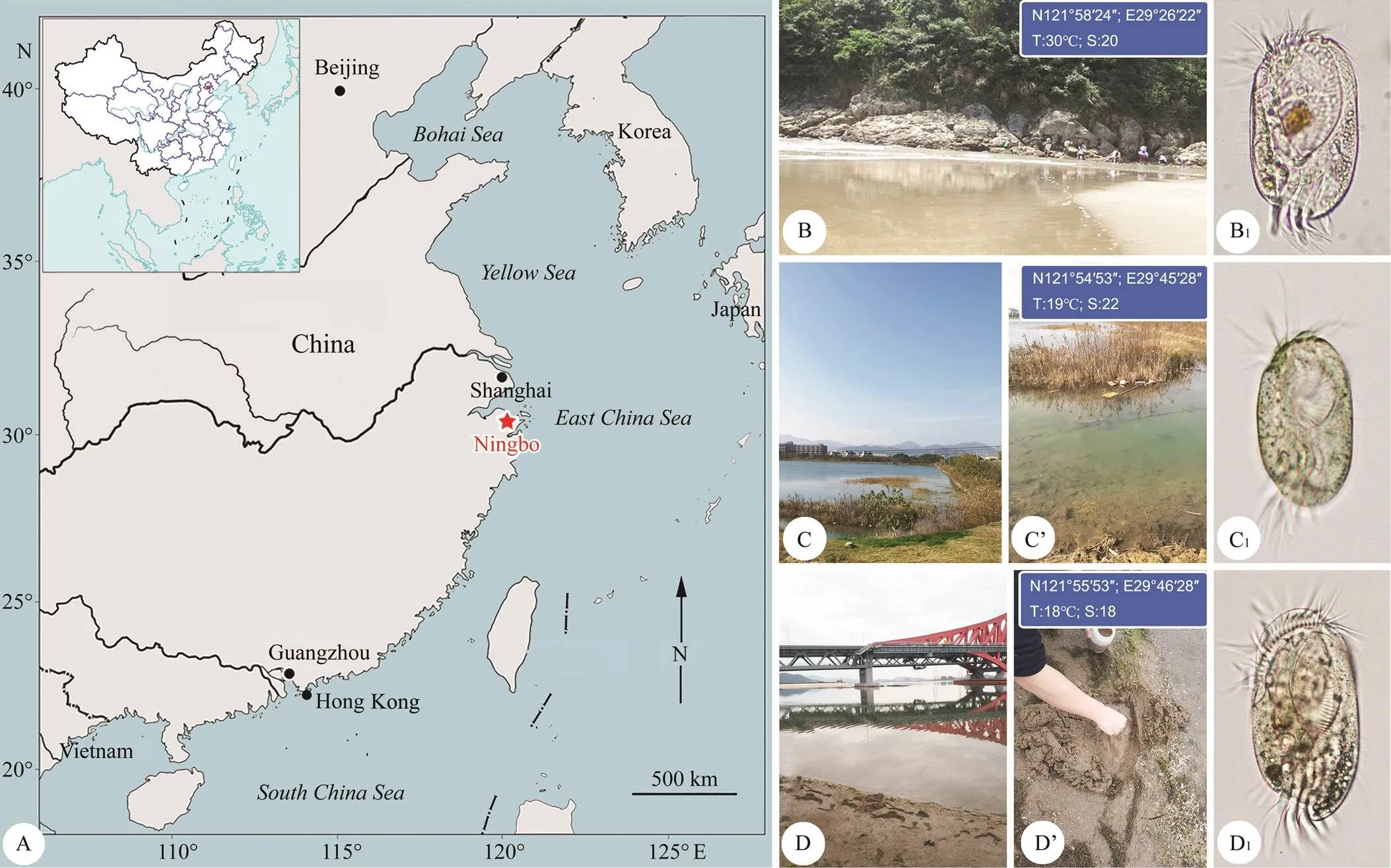
Fig.1 Map and sampling sites. A, Map of the four seas of China, red asterisk indicates the location of Ningbo. B, Coastal beach of Songlanshan at Xiangshan Island. B1, Diophrys quadrinucleata n. sp. collected from site (B). C, C’, A brackish water lake which connected with the Xiangshan Bay, Ningbo. C1, D. oligothrix collected from site (C). D, D’, A sandy beach at Meishan island, Ningbo. D1, D. scutum collected from site (D).
(Fig.1D1) was collected on November 15, 2018 from a sandy beach at Meishan Island, Ningbo (29˚46΄28΄΄E, 121˚55΄53΄΄N), China. The location and ha- bitats of sampling are shown in Figs.1D, D’. The water temperature was about 18℃, salinity was about 18. Samples were taken from intertidal puddles using a sterile sy- ringe.
Specimens were maintained in the laboratory as raw cultures for one to two weeks at room temperature (about 25℃), and rice grains were employed to promote the growth of bacteria as food for the ciliates. Living cells were observed using bright field and differential interference contrast microscopy. The protargol silver staining method from Wilbert (1975) was used to reveal the infraciliature and nuclear apparatus. Counts and measurements of stain- ed specimens were performed at a magnification of 1000×.All drawings were made with the help of a drawing attachment. Terminology and systematics mainly follow Lynn (2008), Song. (2009a) and Gao. (2016).
2.2 DNA Extraction, PCR Amplification and Sequencing
A single cell of each species was isolated from the ori- ginal sample and washed four times with filtered habitat water (0.22µm-pore size membrane, Millipore, USA) and two times with ultra-pure water. Then the cells were trans- ferred to a 1.5mL microfuge tube with a minimum vo- lume of water, respectively. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, CA) according to the manufacturer’s instructions, modified according to Sheng. (2018). PCR amplifications of the SSU rDNA were performed with the primers 18S-F (5’- AACCTGGTTGATCCTGCCAGT-3’) and 18S-R (5’-TG ATCCTTCTGCAGGTTCACCTAC-3’) (Medlin., 1988). Cycling parameters were as follows: initial denaturation of 94℃ for 5min, followed by 18 cycles of amplification (94℃,1min; 66–49℃ touch down, 40s; 72℃, 2min) and another 18 cycles of amplification (94℃, 30s; 48℃, 40s; 72℃, 2min), with a final extension of 72℃ for 7min. The PCR pro-ducts of the new species,and, were sequenced directly in both directions using primers 18S-F, 18S-R and two internal primers (Zhao., 2016; Lian., 2019; Wang., 2019) at GENEWIZ (Beijing, China,incorporated company) and Tsingke Biotechnology (Hang-zhou, China), respectively. Contigs were assembled into one consensus sequence by Seqman (DNAStar).
2.3 Molecular Phylogenetic Analyses
The SSU rDNA sequences of the three newly isolated species and 47 Diophryinaeciliates downloaded from the GenBank database were aligned using MUSCLE v3.7 (Ed-gar, 2004) on the website (URL: http://www.ebi.ac.uk/ Tools/msa/muscle/).,andwere selected as the outgroup taxa. Ambiguous columns were removed with default parameters using Gblock v. 0.91b, resulting in a nucleotide matrix of 1730 sites. Maximum likelihood (ML) analysis, with 1000 boot- strap replicates, was carried out using RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis., 2014) of the CIPRES Science Gateway (URL: http://www.phylo.org/sub_sec- tions/portal). The program MrModeltest v.2.3 (Nylander, 2008) selected the GTR+I+G as the best model with Akaike information criterion (AIC). Bayesian inference (BI) analysis was performed with MrBayes 3.2.6 on XSEDE (Ronquist., 2012), with 1000000 generations, a sam- pling frequency of 100, and a burn-in of 2500 trees. Tree topologies were displayed with MEGA X (Kumar., 2018).
3 Results
3.1 Diophrys quadrinucleata n. sp. (Fig.2, Table 1)
3.1.1 Diagnosis
Cell size (100–140)μm×(60–80)μm; body el- liptical, slightly greyish to yellowish in color; adoral zone comprising about 42 membranelles; five frontal, two ven- tral, five transverse, one left marginal and three caudal cirri; five continuous dorsal kineties with densely arranged dikinetids; four ellipsoidal macronuclear nodules; brackish habitat.
3.1.2 Type locality
Intertidal beach near Songlanshan (29˚26΄22΄΄E, 121˚ 58΄24΄΄N), Xiangshan Island, Ningbo, China. The water temperature was approximately 30℃, and the salinity was about 20.
3.1.3 Etymology
This species has four macronuclear nodules; therefore, its name recalls this feature: quadri-(four), nucleate.
3.1.4 Type material
The slide (registration number: LBR20140823-02-01) with protargol impregnated holotype specimen (Figs.2H, I) is deposited in the Laboratory of Protozoology, Ocean University of China (OUC), China. Two paratype slides (re- gistration numbers: LBR20140823-02-02, 03) are deposited in the Ningbo University.
3.1.5 ZooBank accession number of the new species
Urn:lsid:zooban.org:pub:DDB8B90C-B727-4022-BC74-D1D5E5FACA66.
3.1.6 Deposition of SSU rDNA sequence data
The SSU rDNA sequence is deposited in GenBank with accession number MT109370; its length and GC content are 1736bp and 44.59%, respectively.
3.1.7 Description
Cell size (100–140)μm×(60–80)μm, ratio of length to width about 2:1; body elliptical with both sides straight (Figs.2A–C, F). Dorsoventrally flattened with ven- tral side flat and dorsal side bulging. Apparently sculptured with two longitudinal ribs on ventral side, and obviously depression in central region of body between ribs (Figs.2A, B, G). Anterior end with conspicuous thin, ridged collar and cilia of apical adoral membranelles emerging between ridges (Fig.2J). Posterior end rounded with right lateral concave area on dorsal side where caudal cirri located (Figs.2A, F). Body color slightly greyish to yellowish at low magnification, cytoplasm colorless and transparent, usually packed with numerous granules (about 1– 5μm in diameter) and food vacuoles (containing brow- nish diatoms). Contractile vacuole and cortical granules not recognized. Four macronuclear nodules, spherical to ellipsoidal in shape, each about 20μm×15μm in size, two in a group and diagonally distributed (Figs.2E, H). About four ellipsoidal micronuclei adjacent to macronuclear no- dules (Figs.2E, I). Locomotion by rapid crawling on substrate or freely swimming in water. Feeding on bacteria and diatoms.
Cilia of distal membranelles approximately 25–35μm long. Frontal, ventral and left marginal cirri about 25μm in length. Transverse and caudal cirri very strong with cilia about 40μm and 30μm long, respectively (Figs.2K, L). Dorsal cilia conspicuous, about 8μm long (Figs.2A, F).
Buccal field extending about 2/3 of body length, widest part occupying about 3/5 of body width (Figs.2A, B, F, G). Adoral zone composed of 38–51 membranelles, of which four or five membranelles at distal end reached right side of body; eight to ten membranelles located on dorsal side (Fig.2E). Paroral membrane (PM) curved and long, generally composed of two lines of kinetosomes. Endoral mem- brane (EM) about 3/5 of the length of PM, single-rowed. PM and EM optically intersect near their posterior ends (Figs.2D, H). Five frontal cirri grouped in anterior region of frontal area and always two ventral cirri located together as ‘pre-transverse cirri’. Invariably five strong trans- verse cirri aligned in a row (Figs.2D, H, K); one thin left marginal cirrus positioned behind proximal end of adoral zone of membranelles (Figs.2D, H). Always three caudal cirri at right cell margin with brush-like ends (Figs.2E, I, L). Five dorsal kineties with densely arranged kinetosomes, leftmost and rightmost kineties usually on ventral side (Figs.2E, I).
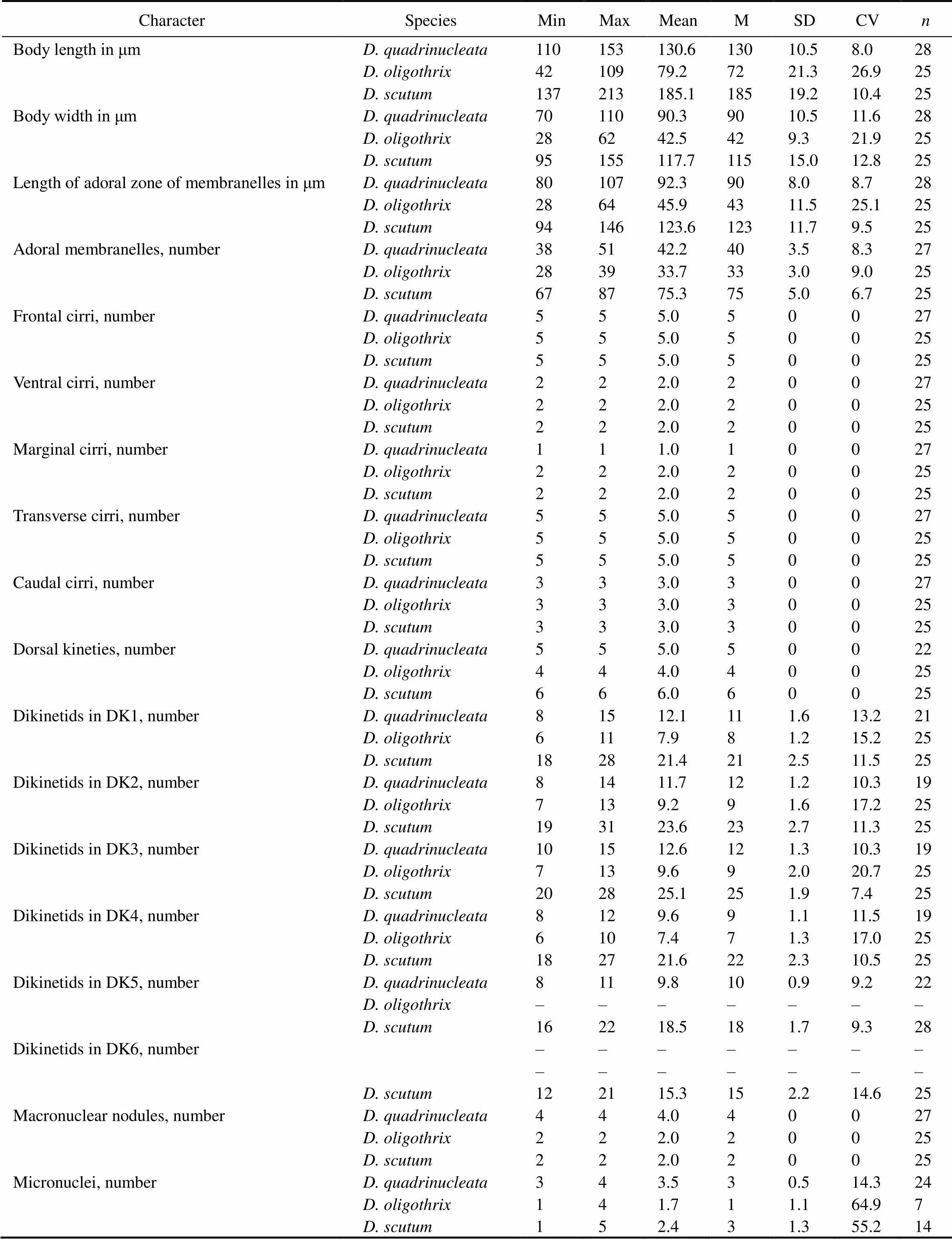
Table 1 Morphometric characterization of Diophrys quadrinucleata n. sp., D. oligothrix, and D. scutum in the present work
Notes: All data are based on protargol-impregnated specimens. CV, coefficient of variation in %; DK1–6, dorsal kinety 1–6; M, median; Max, maximum; Mean, arithmetic mean; Min, minimum;, number of cells measured; SD, standard deviation.
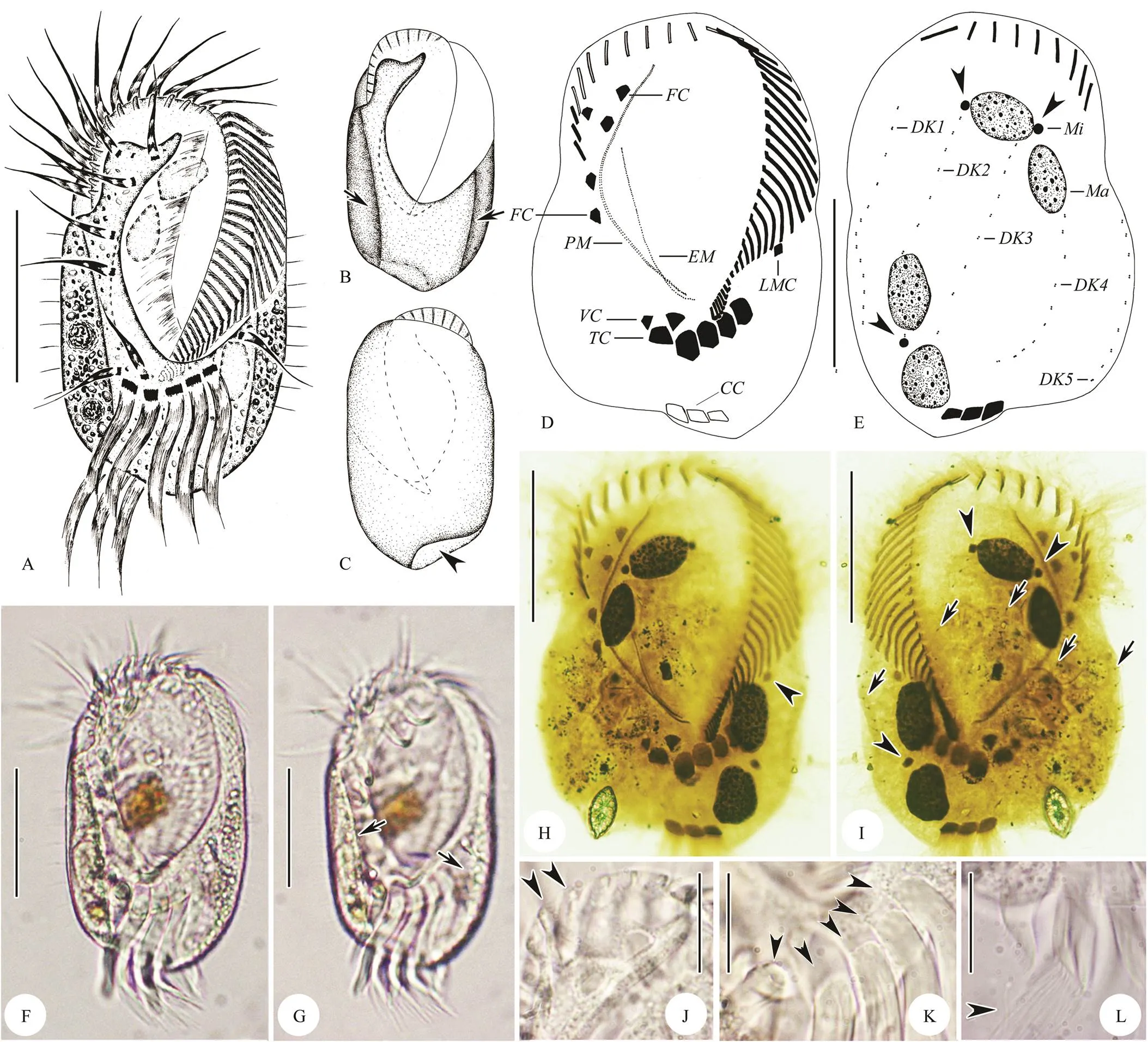
Fig.2 Diophrys quadrinucleata n. sp. from life(A–C, F, G, J–L) and after protargol impregnation (D, E, H, I). A–C, F, G, Ventral and dorsal views of different body shapes and wide oral area, short arrows in (B, G) show two longitudinal ribs on ventral side; arrowhead in (C) indicates the concave area where caudal cirri located. D, E, H, I, Ventral (D, H) and dorsal (E, I) views of the holotype specimen noting infraciliature and nuclear apparatus: arrowheads in (E, I) show the micronucleus; arrowhead in (H) indicates the left marginal cirrus; short arrows in (I) show densely arranged dikinetids within dor- sal kineties. J–L, Amplification of some structures: arrowheads in (J) mark the conspicuous and ridged collar, arrowheads in (K) showing the five strong transverse cirri; arrowhead in (L) shows the brush-like end of caudal cirri. CC, caudal cirri; DK1–5, dorsal kinety 1–5; EM; endoral membrane; FC, frontal cirri; LMC, left marginal cirrus; Ma, macronucleus; Mi, micronuclei; PM, paroral membrane; TC, transverse cirri; VC, ventral cirri. Scale bars=50μm (A–I), 20μm (J–L).
3.2 Diophrys oligothrix Borror, 1965 (Figs.3, 4, Table 1)
3.2.1 Description of Ningbo population
Cell size (60–80)μm×(40–45)μm, body shape elliptical to oval with left side nearly straight and right margin convex (Figs.3A, F, G, 4A). Anterior end with con- spicuous ridged collar accompanied by ridges, cilia of api- cal adoral membranelles emerge near these ridges (Fig.3A). Posterior end rounded with right concave area on dorsal side where caudal cirri located. Bright greenish spherical cortical granules approximately 0.5μm across, on dorsal surface, densely arranged in irregular lines and about seven to nine granules packed together around every dorsal cil-ium in a rosette-pattern (Fig.3E, arrowheads, 4D, circles); on ventral surface, packed around the cirri and along the right edge of oral area (Figs.3C, D, 4C). Cytoplasm colorless and transparent, usually packed with numerous gra- nules (3–7.5μm in diameter) and food vacuoles (containing diatoms, some big diatom almost as long as body length) (Fig.4D). Contractile vacuole not recognized. Always two sausage-shaped macronuclear nodules with each one located in anterior and posterior half of cell (Figs.3I, 4F). One to four spherical or ellipsoidal micronuclei adjacent to macronuclear nodules (Figs.3I, 4H). Locomotion by rapid crawling on substrate or freely swimming in water. Feeding on bacteria and diatoms.
Cilia of distal adoral membranelles approximately 25μm long. Frontal, ventral and marginal cirri thin and about 20μm in length. Transverse and caudal cirri very strong, 30μm and 20μm long, respectively. Dorsal cilia about 6μm long and obviously detectable.
Buccal field extending about 1/2 of body length andadoral zone composed of 28–39 membranelles. Paroral membbrane (PM) and endoral membrane (EM) optically intersecting near their posterior ends. PM almost the same length as buccal field, slightly curved, extending near the first frontal ventral cirrus, EM about 3/5 of PM in length. Five frontal cirri grouped in frontal area of cell and always two ventral cirri located together as ‘pre-transverse cirri’ (Fig.4F). Invariably five strong transverse cirri aligned in a row; three caudal cirri at right cell margin. Two left marginal cirri, of which one beneath proximal end of AZM, the other near the leftmost transverse cirrus. Four dorsal kineties with loosely arranged dikinetids. (Figs.3H, I, 4G).

Fig.3 Diophrys oligothrix from life(A–G) and after protargol impregnation (H, I). A, F, G, Ventral (A, F) and dorsal (G) views to show different body shapes. B, The lateral view showing the sculptured body margin. C–E, Ventral (C, D) and dorsal (E) views to show numerous tiny cortical granuleswhich distribute around the cirri and along the paroral membrane on the ventral surface (C, D), arranging in irregular lines and around every dorsal cilium in a rosette-pattern on the dorsal side (arrowheads in E to show the granules around the dorsal cilium). H, I, Ventral (H) and dorsal (I) views of a typical specimen noting the infraciliature and nuclear apparatus: arrowheads in (H) indicate two left marginal cirri. CC, caudal cirri; DK1–4, dorsal kinety 1–4; EM, endoral membrane; FC, frontal cirri; Ma, macronucleus; Mi, micronuclei; PM, paro- ral membrane; TC, transverse cirri; VC, ventral cirri. Scale bars=30μm.
3.2.2 Deposition of SSU rDNA sequence data
The SSU rDNA sequence is deposited in GenBank with accession number MT109371. Its size and GC content are 1656bp and 44.32% respectively.
3.2.3 Ecology and distribution
Ningbo population ofwas collect- ed on 15 November 2018. Water temperature was 19℃, sa- linity was about 22.
New Hampshire, USA (Borror, 1965b; tidal marsh pools); Coastal of UK (Carey, 1992, marine interstitial area; Hu., 2012, a salt marsh near Blakeney village); Qingdao, China (Song and Wilbert, 1994, seawater ponds; Song andPackroff, 1997, coastal waters; Chen and Song, 2002, shrimp and shellfish farming ponds; Song., 2009a, coastal waters); King George Island, Antarctica (Song and Wilbert, 2002, seawater ponds); Guangdong, China (Chen and Song, 2002, coastal waters of Zhanjiang); Nagasaki, Japan (Luo., 2014, a fishing port).

Fig.4 Diophrys oligothrix from life (A, D from bright field; B, C, E from differential interference contrast microscopy) and after protargol impregnation (F–H). A, Ventral view of a representative individual. B, Ventral view of a slightly squashed specimen: arrowheads to show two widely separated left marginal cirri. C, D, Ventral (C) and dorsal (D) views to show numerous tiny yellow-greenish cortical granules: arrowheads in (C) indicate the cortical granules distributed around the cirri and along the right edge of oral area; circles in (D) to show the cortical granules arranged irregular lines and around every dorsal cilium. E, Lateral view showing the sculptured body margin. F, G, Ventral (F) and dorsal (G) views of a typical specimen showing infraciliature and nuclear apparatus: arrowheads in (F) indicate two left marginal cirri; arrowheads in (G) show regularly arranged dikinetids within dorsal kineties. H, Detail of nuclear apparatus: arrowhead in (H) show micronuclei. Scale bars=30μm.
3.3 Diophrys scutum (Dujardin, 1841) Kahl, 1932 (Fig.5, Table 1)
3.3.1 Description of Ningbo population
Elliptical cell (150–170)μm×(70–85)μm(Figs. 5A–C, F). Ventral side sculptured with two longitudinal ribs (Fig.5B). Anterior end with conspicuous, large collar; posterior end lightly pointed with right concave area on dorsal side where caudal cirri located (Figs.5A, F). Cytoplasm colorless and transparent, packed with granules (3– 8.5μm in diameter) and food vacuoles (Figs.5F, G). Contractile vacuole not recognized. Dorsal side densely covered with symbiotic bacteria/archaeobacteria (?) (Fig.5J). Two sausage-shaped macronuclear nodules (Figs.5E, H). One to five spherical micronuclei adjacent to macronuclear nodules (Fig.5H, arrows).
Buccal field extending about 2/3 of body length. Adoral zone composed of 67–87 membranelles. Paroral membrane (PM) and endoral membrane (EM) optically intersecting near middle part of EM. PM long, slightly curved, extend- ing almost to anterior end of cell, while EM about three quarters of PM in length (Figs.5D, H). Five frontal cirri approximately in a line along anterior right margin, two strong and developed ventral cirri located together as ‘pre- transverse cirri’; five strong transverse and three caudal cirri; two thin left marginal cirri widely separated (Figs. 5H, K). Six dorsal kineties with densely distributed dikinetids on dorsal and lateral ventral sides (Figs.5E, I).
3.3.2 Deposition of SSU rDNA sequence data
The SSU rDNA sequence is deposited in GenBank with accession number MT109372; its size and GC content are 1669bp and 44.46% respectively.
3.3.3 Ecology and distribution
The Ningbo population ofwas collected on November 15, 2018. Water temperature was 18℃, salinity was about 18.
New Hampshire, USA (Borror, 1965b, tidal marsh pools); Japan Sea (Raikov and Kovaleva, 1968, sandy sediments); Caspian Sea (Agamaliev, 1971, western coast); UK (Carey, 1992, marine interstitial area); Qingdao, China (Song and Packroff, 1997, coastal waters; Chen and Song, 2002, shrimp farming pond; Song., 2009a, coastal waters).
3.4 SSU rDNA Sequence and Phylogenetic Analyses (Fig.6)
The BI and ML trees inferred from SSU rDNA sequences had similar topologies for the-complex group regardless of the number of taxa included in the preliminary analyses. As shown in Fig.6,n. sp. grouped withandin both methods while the support values are va- riable (53% ML, 0.96 BI). Our population offalls within a big cluster ofsequences (99% ML, 1.00 BI). In contrast, species ofdo not form a monophyletic lineage, as sequences under the names ofcf.,(JN172996),(MG603601) are nested within it. How- ever, their internal relationships are far from robust as in- dicated by the poor support values. The newly isolated po- pulation ofgrouped well with a British po- pulation (86% ML, 1.00 BI), followed by a sequence under the name of(MG603601), although this grouping only receives very low support (21% ML, 0.55 BI). The other two sequences ofclustered with, forming the sister clade to the big cluster ofand several congeners.

Fig.5 Diophrys scutum from life(A–C, F, G, J from bright field; K from differential interference contrast microscopy) and after protargol impregnation (D, E, H, I). A–C, F, G, Ventral (A, B, F) and dorsal (C, G) views to show different body shapes: arrowheads in (G) show numerous granules. D, E, H, I, Ventral (D, H) and dorsal (E, I) views of a typical specimen showing infraciliature and sausage-shaped nuclear apparatus: arrowheads in (D, H) indicate the two left marginal cirri; arrowheads in (I) show densely arranged dikinetids within dorsal kineties; arrows in (H) to show micronuclei. J, Dorsal side of slightly squashed specimen: arrowheads to show the densely distributed symbiotic bacteria/archaeobacteria (?). K, Ventral view showing strong transverse cirri: arrowheads indicate two left marginal cirri; short arrows denote two ventral cirri. CC, caudal cirri; DK1–6, dorsal kinety 1–6; EM, endoral membrane; FC, frontal cirri; Ma, macronucleus; PM, paroral membrane; TC, transverse cirri; VC, ventral cirri. Scale bars=50μm (A, D, E, F–I), 20μm (J, K).

Fig.6 The maximum-likelihood (ML) tree inferred from the small subunit rDNA sequences of 50 species in the family Uronychiidae, showing the positions of Diophrys quadrinucleata n. sp., D. oligothrix and D. scutum. Numbers near nodes are non-parametric bootstrap values for ML and posterior probability values for Bayesian inference (BI). Solid circles represent 100% for ML and 1.00 for BI. The scale bar corresponds to 1 substitution per 100 nucleotide positions. All branches are drawn to scale.
4 Discussion
The genusis characterized by having stable front-ventral cirri, that is, five front and two ventral cirri; one or two left marginal cirri near the proximal portion of the adoral zone, two separated undulating membranes and three posterior-laterally located caudal cirri (Hu., 2012). Till now, only ten nominal species mainly from marine habitat have been reported. Two species of them lack infraciliature and/or molecular data, namelyRuinen, 1938 andBorror, 1965. Based on the previous researches and present findings, ninemorphospecies with infraciliature are currently well recognized:n. sp.;Song., 2009a;(Ehrenberg, 1838) Sche- wiakoff, 1893;Hu., 2012;Hu, 2008;Borror, 1965;Shen., 2011;Luo., 2014;(Dujardin, 1841) Kahl, 1932. Detailed comparisons of morphological characteristics are listed in Table 2. We here supply a key for the identification of nine well- described species (for illustrations of selected key characters, see Fig.7).
The key for the identification of nine well-described species


Table 2 Morphometrical and morphological comparison of nine related and well-described Diophrys species
Notes: AZM, adoral zone of membranelles; DK, dorsal kineties; LMC, left marginal cirri; Ma, macronucleus; * This character is not shown in figures, but described in the text. References: [1] present work; [2] Song and Packroff (1997); [3] Hu. (2012); [4] Luo. (2014); [5] Chen and Song (2002); [6] Song. (2009a); [7] Hu (2008); [8] Fan. (2013); [9] Shen. (2011).
4.1 Diophrys quadrinucleata n. sp.
Before the present work, most of knownspecies have two ellipsoidal or sausage-shaped macronuclear nodules. Hu. (2012) described a multi-macronucleus species,, which has 7–23 macronuclear nodules. It cannot be confused withn. sp. according to the character of macronucleus. Additionally,can be distinguished from the new species by the number of left marginal cirri (one. two). Kattar (1970) reported awith four macronuclear nodules and named it as. Except for the number of macronuclear nodules, it is very similar toin other morphological features. Indeed the validity of this species was even doubted by the original author. We agree with Song. (2007) thatis a special population of. In fact, it can be easily distinguished fromn. sp. by the number of left marginal cirri (two. one in new species).
Althoughis a rare species that has never been described with infraciliature and molecular information since it was originally reported by Ruinen (1938), it can be well distinguished from the new species by some characteristics, such as smaller body size (30–40μm. 100–140μm), well-developed anterior adoral membranelles (. normal frontal adoral membranelles size), the short and fine frontal-ventral-transverse cirri (. normal frontal- ventral cirri size, developed and strong transverse cirri), and the absence of the left marginal cirri (. presence).
The information ofis also incomplete and it has never been redescribed since the original description by Borror (1965a). However, this understudied species can also be clearly distinguished fromn. sp. by having two left marginal cirri (. single left marginal cirrus), two macronuclear nodules (. four), and eight dorsal kineties (. five).
The remaining well-described eight species can be divided into two groups based on the number of left marginal cirri.,,,,andhave two left marginal cirri, whileandonly possess single left marginal cirrus (Fig.7). Our new speciesn. sp. can be easily dis- tinguished from the majority of its congeners by having single left marginal cirrus. Thus, only two species with single left marginal cirrus need to be compared with. How- ever, the new species clearly differs fromandas it has four spherical to ellipsoidal ma- cronuclear nodules (. two ellipsoidal nodules). Moreover, the distinctions betweenn. sp. and its two congeners are also demonstrated by the divergence of the SSU rDNA sequences. There are seven and 20 nucleotide differences between the new species andand, respectively. Therefore,n. sp. is identified as a well-outlined and distinctive member of the genus.

Fig.7 Illustrated key to nine Diophrys species with infraciliature information. AZM, adoral zone of membranelles; DK, dorsal kinety; Ma, macronucleus. Scale bars=50μm.
4.2 Diophrys oligothrix Borror, 1965
was discovered by Borror (1965b) in a tidal marsh pool in New Hampshire, USA. In the following half century, it was isolated and identified from a variety of habitats all over the world, such as marineinterstitial area or salt marsh in UK, coastal waters orfarming ponds in China, seawater ponds in Antarctica, and fishing port in Japan (Carey, 1992; Song and Wilbert, 1994, 2002; Song and Packroff, 1997; Chen and Song, 2002; Hu., 2012; Luo., 2014). Our population isolated from the brackish-water lake corresponds well with previously published populations in terms of living morphology and ciliary pattern, especially the continuous dorsal kineties with loosely arranged cilia. The slightly dis- similar number of adoral membranelles between the Ning- bo population and the original description was considered as a difference between populations (Curds and Wu, 1983; Song., 2007). Thus, the Ningbo population was iden- tified as.
In the phylogenetic trees (Fig.6), the Ningbo population grouped well with the Blakeney population from UK (JN172995, Hu., 2012), supporting the morphological identification of it as a population of. How- ever, these two populations did not form a monophyletic clade with the Qingdao population isolated from China (DQ353850, Song., 2009a). The above three populations cannot be further distinguished from each other based on morphological characters, although their sequence di- vergences of the SSU rDNA range from 11 to 18 nucleotide sites (Table 3). Thus, it is reasonable to deduce that there might be cryptic species in, which need further information to distinguish and define them.

Table 3 SSU rDNA sequence similarities between three Diophrys oligothrix populations and D. blakeneyensis
4.3 Diophrys scutum (Dujardin, 1841) Kahl, 1932
is the largest species in the genus and has been reported several times (Borror, 1965b; Agamaliev, 1971; Song and Packroff, 1997; Song., 2007). The detailed morphological characteristics were provided based on a Qingdao population in Song and Packroff (1997). Our population resembles with the previous po- pulations very well in both living morphology and ciliary patterns. The only difference is that we found densely dis- tributed symbiotic bacteria/archaeobacteria (?) on the dor- sal surface of the Ningbo population, which was never men- tioned previously (Agamaliev, 1971; Song and Packroff, 1997). The symbiotic bacteria/archaeobacteria (?) covering the dorsal surface are very similar to cortical granules in the living normal species. However, these particles can be easily separated from the dorsal surface in the slightly squeezed cells. Recently, Luo. (2014) described a Ja- panese population ofcf.with numerous tiny, colourless, rice-shaped cortical granules densely ar- ranged on dorsal side. It is highly possible that these particles are symbiotic bacteria/archaeobacteria (?) and are misinterpreted as cortical granules. Furthermore, all se-quences ofform a well-supported mono- phyletic cluster in the present phylogenetic trees and the sequence divergences of the SSU rDNA are less than four nucleotide sites each other, justifying the morphological identification of this organism.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31970398), the National Key Research and Development Program of China (No. 2018YFD0900701), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (No. 2019 333), the Technology Innovation Team of Ningbo City (No. 2015C110018), and the K. C. Wong Magna Fund in Ning- bo University.
Agamaliev, F. G., 1971. Complements to the fauna of psammophilic ciliates of the Western coast of the Caspian Sea., 8: 379-404 (in Russian with English summary).
Borror, A. C., 1965a. Morphological comparison of(Dujardin, 1841) andn. sp. (Hypotrichi- da, Ciliophora)., 12: 60-66.
Borror, A. C., 1965b. New and little-known tidal marsh ciliates., 84: 550- 565.
Carey, P. G., 1992.. Chapman & Hall, London, 351pp.
Chen, Z. G., and Song, W. B., 2002. Characterization and identification of thespecies (Protozoa, Ciliophora, Hypo- trichida) based on RAPD fingerprinting and ARDRA riboprinting.,38: 383-391.
Curds, C. R., and Wu, I. C. H., 1983. A review of the Euplotidae (Hypotrichida, Ciliophora)., 44: 191-247.
Dujardin, F., 1841. Histoire naturelle des zoophytes. Infusoires, comprénant la physiologie et la classification de ces animaux, et la manière de les étudier al’aide du microscope., Paris.
Edgar, R. C., 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput., 32: 1792-1797.
Fan, Y. B., Warren, A., Al-Rasheid, K. A. S., Chen, X., and Shao, C., 2013. Morphology and SSU rRNA gene-based phylogeny of two-like ciliates from northern China, with notes on morphogenesis of(Protozoa, Ci- liophora)., 274: 395-403.
Gao, F., Warren, A., Zhang, Q. Q., Gong, J., Miao, M., Sun, P., Xu, D. P., Huang, J., Yi, Z. Z., and Song, W. B., 2016. The all- data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata)., 6: 24874.
Hill, B. F., and Borror, A. C., 1992. Redefinition of the generaandand establishment of the genusn. g. (Ciliophora, Hypotrichida): Implication for the species problem., 39: 144-153.
Hu, X. Z., 2008. Cortical structure in non-dividing and dividingspec. nov. (Ciliophora, Euplotida) with noteson morphological variation.,44: 115-129.
Hu, X. Z., Huang, J., and Warren, A., 2012. The morphology and phylogeny of two euplotid ciliates,spec. nov. andBorror, 1965 (Protozoa, Ciliophora, Euplotida)., 62: 2757-2773.
Hu, X. Z., Lin, X. F., and Song, W. B., 2019.Science Press, Beijing, 936pp.
Huang, J., Dunthorn, M., and Song, W. B., 2012. Expanding character sampling for the molecular phylogeny of euplotid ciliates (Protozoa, Ciliophora) using three markers, with a fo- cus on the family Uronychiidae., 63: 598-605.
Jankowski, A. W., 1979. Revision of the order Hypotrichida Stein, 1859. Generic catalogue, phylogeny, taxonomy., 86: 48-85.
Jankowski, A. W., 2007. Phylum Ciliophora Doflein, 1901. In:.Alimov, A. F., ed.,St. Petersburg, Nauka, 415-993.
Jiang, J. M., and Song, W. B., 2010. Two new-like genera and their type species,n. g., n. sp. andn. g., n. sp. (Ciliophora: Euplotida), with notes on their molecular phylogeny., 57: 354-361.
Jiang, J. M., Warren, A., and Song, W. B., 2011. Morphology and molecular phylogeny of two new marine euplotids,n. g., n. sp., andn. sp. (Ciliophora: Euplotida)., 58: 437-445.
Kahl, A., 1932. Urtiere oder Protozoa I: Wimpertiere oder Ci- liata (Infusoria). 3. Spirotricha., 25: 399-650.
Kattar, M. R., 1970. Estudo dos protozoarios ciliados psamo- filos do litoral Brasileiro., 27: 123-206.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Mentjies, P., and Drummond, A., 2012. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data., 28: 1647-1649.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., 2018. Mega X: Molecular evolutionary genetics analysis across com- puting platforms., 35: 1547-1549.
Lian, C. Y., Zhang, T. T., Al-Rasheid, K. A. S., Yu, Y. H., Jiang, J. M., and Huang, J., 2019. Morphology and SSU rDNA- based phylogeny of twospecies from China:sp. n. andKahl, 1932 (Ciliophora, Euplotida)., 6: 1-14.
Luo, X. T., Hu, X. Z., and Suzuki, T., 2014. Microscopic investigation of three species of(Ciliophora, Euplotida, Uronychiidae) from Japan, includingnov. spec., 50: 496-508.
Lynn, D. H., 2008.. 3rd edition. Springer Press, Dordrecht, 605pp.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L., 1988. The characterization of enzymatically amplified eukaryotic 16S- like rRNA-coding regions., 71: 491-499.
Nylander, J. A. A., 2008. MrModeltest 2.3. Department of Systematic Zoology, Uppsala University, Uppsala, Sweden.
Raikov, I. B., and Kovaleva, V. G., 1968. Complements to the fauna of psammobiotic ciliates of the Japan Sea (Posjet Gulf)., 6: 309-333.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M. A., andHuelsenbeck, J. P., 2012. MrBayes 3.2: Efficient Bayesian phy- logenetic inference and model choice across a large model space., 61: 539-542.
Ruinen, J., 1938. Notizen über Ciliaten aus konzentrierten Salzgewässern., 20: 243-256.
Shen, Z., Yi, Z. Z., and Warren, A., 2011. The morphology, on- togeny, and small subunit rRNA gene sequence analysis ofn. sp, (Protozoa, Ciliophora, Eu- plotida), a new marine ciliate from coastal waters of southern China., 58: 242-248.
Sheng, Y. L., He, M., Zhao, F. Q., Shao, C., and Miao, M., 2018. Phylogenetic relationship analyses of complicated class Spi- rotrichea based on transcriptomes from three diverse micro- bial eukaryotes:,and., 129: 338-345.
Song, W. B., and Packroff, G., 1997. Taxonomische Untersu- chungen an marinen Ciliaten aus China mit Beschreibungen von zwei neuen Arten,nov. spec. undnov. spec. (Protozoa, Ciliophora)., 147: 331-360.
Song, W. B., and Wilbert, N., 1994. Morphogenesis of the marine ciliateBorror, 1965 during the cell division (Protozoa, Ciliophora, Hypotrichida)., 30: 38-44.
Song, W. B., and Wilbert, N., 2002. Faunistic studies on marine ciliates from the Antarctic benthic area, including descriptions of one epizoic form, 6 new species and 2 new genera (Protozoa: Ciliophora)., 41: 23-61.
Song, W. B., Shao, C., Yi, Z. Z., Li, L. Q., Warren, A., Al- Rasheid, K. A. S., and Yang, J. P., 2009a. The morphology, mor- phogenesis and SSU rRNA gene sequence of a new marine ciliate,spec. nov. (Ciliophora; Euploti- da)., 45: 38-50.
Song, W. B., Warren, A., and Hu, X. Z., 2009b.Science Press, Bei- jing, 518pp.
Song, W. B., Wilbert, N., Al-Rasheid, K. A. S., Warren, A., Shao, C., Long, H. A., Yi, Z. Z., and Li, L. Q., 2007. Redescriptions of two marine hypotrichous ciliates,and(Ciliophora, Euplotida), with a brief revision of the ge- nus., 54: 283- 296.
Stamatakis, A., 2014. RAxML version 8: A tool for phylogene- tic analysis and post-analysis of large phylogenies., 30: 1312-1313.
Wang, Y. R., Wang, C. D., Jiang, Y. H., Katz, L. A., Gao, F., andYan, Y., 2019. Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny., 62:203-214.
Wilbert, N., 1975. Eine verbesserte Technik der Protargolimpräg- nation für Ciliaten., 64: 171-179.
Zhao, Y., Yi, Z. Z., Gentekaki, E., Zhan, A., Al-Farraj, S. A., and Song, W. B., 2016. Utility of combining morphological char- acters, nuclear and mitochondrial genes: An attempt to re- solve the conflicts of species identification for ciliated protists., 94: 718-729.
#The two authors contributed equally to this work.
. E-mail: xiangruichen@126.com
March 17, 2020;
April 13, 2020;
June 4, 2020
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Centurial Evolution of an Offshore Mud Deposition Area in the Changjiang (Yangtze) Estuary and Its Links to Environmental and Anthropogenic Activities
- Characteristics and Origins of Suspended Pyrite in the Mixing Zone of the Yangtze Estuary
- Characterization of Fe(III)-Reducing Enrichment Cultures and Isolation of Enterobacter sp. Nan-1 from the Deep-Sea Sediment, South China Sea
- Evolution of Palaeoenvironment of the South Yellow Sea Since the Last Deglaciation
- Sulfate-Methane Transition Depths and Its Implication for Gas Hydrate
- Comprehensive Investigation and Assessment of Nutrient and Heavy Metal Contamination in the Surface Water of Coastal Bohai Sea in China
