Inhibition of ε-Poly-lysine on Dominant Spoilage Bacteria in Grass Carp and Its Effect on Microbial Diversity during Cold Storage
2020-09-21HOUWenfuYUEQiqiHANQianhuiWANGHongxunZHOUMinMINTingPANSiyi
HOU Wenfu, YUE Qiqi, HAN Qianhui, WANG Hongxun, ZHOU Min, MIN Ting, PAN Siyi
(1. School of Food Science and Engineering, Wuhan Polytechnic University, Wuhan 430023, China; 2. College of Food Science and Technology, Huazhong Agricultural University, Wuhan 430070, China; 3. School of Biology and Pharmaceutical Engineering,Wuhan Polytechnic University, Wuhan 430023, China)
Abstract: The present study investigated the effect of ε-poly-lysine (ε-PL) pretreatment on spoilage bacteria growth in grass carp (Ctenopharyngodon idellus) fillets under refrigerated storage as well as on its microbial diversity using 16S rRNA high-throughput sequencing technique. Results showed that 1.0 mg/mL ε-PL could inhibit the growth of Pseudomonas sp.,Shewanella sp. and Aeromonas sp., which were isolated and identified as the dominant spoilage bacteria from grass carp fillets. A total of 858 964 bacterial sequences targeting the V3-V4 region of 16S rRNA and 388 different genera belonging to 28 phyla were observed by high-throughput sequencing. The microbial diversity of both control and ε-PL pretreated groups decreased as the storage time increased. Pseudomonas sp. was the absolute dominant spoilage bacterium in fresh grass carp fillets. ε-PL pretreatment affected both the microbial diversity and abundance. Notably, it had a very significant influence on Flavobacterium in fresh grass carp fillets (P < 0.01), and a significant influence on Pseudomonas in 6-day-stored samples(P < 0.05). In conclusion, ε-PL presents a high performance in inhibiting the growth of fish spoilage bacteria and can lead to changes in the structure of the microbiome in grass carp fillets, which supports its application in fresh fish preservation.
Keywords: grass carp; ε-poly-lysine; high-throughput sequencing; microbial diversity
ε-Poly-lysine (ε-PL) is one kind of widely used natural antibacterial agents because of the good efficiency and broad antimicrobial-spectrum, which consists of 25-30L-lysine residues linked by peptide bonds betweenε-amino andα-carboxyl groups[1]. Moreover,ε-PL has been recognized as safe food additives by Food and Drug Administration (FDA,USA) and Chinese Food and Drug Administration (CFDA,China). Currently, it has been used in many food products to prolong the shelf life, such as cheese, salad-dressings, fish and sauces, etc.[2]. Compared with some chemical additives,ε-PL presented water solubility, edibility, biodegradability and nontoxicity towards humans and the environment, and without showing any side effects[3].
Grass carp (Ctenopharyngodon idellus) is one of the most important and favored commercial freshwater fish species, which accounts for its high nutritional values rich in proteins, vitamins, and minerals[4]. The aquaculture production of grass carp reached 5 537 794 tons per year in the worldwide[5]. However, fresh fish are easy to decompose because of protein degradation, lipid oxidation or decomposition caused by the spoilage microorganisms[6].Hence, extending the preservation time of fresh fish is an urgent issue. Researches demonstrated that some methods presented high performance in prolonging the shelf life of grass carp, such as salting, adding polyphenols, and coating with chitosan[7-9]. But there are few report that investigated the effect ofε-PL on the growth of fish-spoilage bacteria, and howε-PL affect the microbial composition of grass carp[10].
Some antimicrobials could selectively inhibit the growth of certain bacteria. The major bacteria in microbial communities determine the various spoilage phenomena[11].High-throughput sequencing (HTS) have gradually been applied in foods to assess the changes in microbial populations during food production and storage[12-15]. de Filippis et al[16]used HTS of 16S rRNA gene amplicons to explore the microbial diversity in beefsteaks under cryopreservation and to investigate the contamination variability in its processing. In-depth study of bacterial community and its dynamics evolution will help to improve the understanding of spoilage progress and provide a basis for selection of antimicrobial agents.
The aim of the present study was to assess the effect ofε-PL pretreatment on the growth of spoilage bacteria isolated from fish in term of minimum inhibitory concentration (MIC)and HTS was used to characterize the changes in bacterial communities of grass carp (Ctenopharyngodon idellus) fillets treated withε-PL during refrigerated storage, with a hope to support the application ofε-PL in fresh fish preservation.
1 Materials and Methods
1.1 Materials and reagents
The 10-tail of fresh grass carp (Ctenopharyngodon idellus) with an average body mass of (1 500 ± 50) g were purchased from the local fisheries market in Wuhan, China.
Sodium chloride Sinopharm Chemical Reagent Co.,Ltd.; plate count agar (PAC), thiosulfate citrate bile salts sucrose agar (TCBS), aeromonas medium base (RYAN),PseudomonasCFC selective agar Qingdao Hope Bio-Technology Co., Ltd.;ε-poly-lysine (food grade)Zhengzhou Bainafo Biological Engineering Co., Ltd..
1.2 Instrumentations
HBM-400D homogenizer Tianjin Hengao Company; DNA extraction Kit American Omega B i o-Te k C o m p a n y; N a n o D r o p 2 0 0 0 U V-V I S Spectrophotometer, Quanti Fluor™-ST fluorimeter American Thermo Fisher Scientific Inc.; Bioanalyzer 2100 system American Agilent Company.
1.3 Methods
1.3.1 Sample preparation
After stunning the grass carp, they were cleaned and cut into pieces of uniform size (body mass (25 ± 5) g) with a sterilized knife. The moisture on the surface of the fish was removed by a sterile filter paper. Each set of the fresh grass carp was randomly divided into 28 sample boxes, including the control (12 boxes),ε-PL treated (12 boxes) and 4 boxes for backup. The control group received no treatment and the pre-treatment group withε-PL was operated by immersing fish block into 1.0 mg/mLε-PL aqueous solution with a volume ratio of 1:3 (m/V) for 5 min. After pre-treatment, all samples were put into the sterile sample box separately and placed in the refrigerator at 4 ℃ for 0, 3, 6, 9 days until the grass carp fillets was thoroughly corrupt. The 0-day storage of controlled group was regarded as blank measurement.
1.3.2 Bacteria isolation and identification
After being stored for 6 days under refrigerated storage (4 ± 1) ℃, the conteolled group sample of fish meat was aseptically weighed and then was transferred to a stomacher bag with 225 mL sterile 0.85% NaCl and mixed homogenously for 30 s using a stomacher. A loop of the culture was then steaked onto the surface of PCA medium.After incubation at its optimum temperatures (30 ℃) for 48 h, typical single colonies on medium were selected and observed. All colonies on the PCA plate were selected,inoculated and cultured in lysogeny broth (LB) medium for 24-36 hours, and then repeatedly separated by scribing on the PCA plate until single colonies were obtained. After observing their morphological characteristics, the strains were identified corresponding to 16S rRNA.
1.3.3 Sample treatment
The MIC ofε-PL on fish-spoilage bacteria was determined using the standard broth microdilution method with some modifications[17].Pseudomonassp. was incubated at 30 ℃,Shewanellasp. andAeromonassp. were incubated at 25℃ for 8-10 h to approximately 106-107CFU/mL in LB medium. Serial dilutions ofε-PL were prepared to obtain final concentrations of 0.031 25, 0.062 5, 0.125, 0.25, 0.5, 1, 2,4 mg/mL in LB medium. Simultaneously, 10 mL of bacterial suspension was defined as negative controlled group, and LB broth was set as blank controlled group. The tubes were incubated at different temperature (Pseudomonassp. at 30 ℃,Shewanellasp. andAeromonassp. at 25 ℃) for up to 24 h before recording the MICs. The lowest concentration ofε-PL without any bacteria growth was defined as MIC.
1.3.4 Measurement of accounting of total viable colony andPseudomonassp.
The fresh grass carp meat was divided into 5 groups.Each group of grass carp meat was immersed in 0, 0.5, 1.0,2.5, 5.0 mg/mLε-PL respectively for 5 min, and the group immersed in sterile water (0 mg/mLε-PL) was regarded as the control group. Subsequently, all samples were packaged and kept at (4 ± 1) ℃ for subsequently bacterial analysis.The total number of bacteria colonies was detected every other day. Briefly, samples (0.1 mL) that prepared by homogenization according to section 1.3.2 description were diluted 10 times with 0.85% NaCl, spread on the surface of PCA andPseudomonasCFC Selective Agar and incubated at (30 ± 1) ℃ for 48 h. All tests repeated in triplicate and the unit of colonies was recorded as lg (CFU/g).
1.3.5 DNA extraction and PCR amplification
Bacterial DNA extracted fromε-PL treated samples and controlled group on Day 0, 3, 6, 9 by sampling on the surface using a cotton swab, according to the method described by Huang Zhan et al[18]. DNA was extracted using a DNA extraction Kit according to the manufacturer’s instructions. The concentration and quality of the extracted DNA were measured using a Nano Drop 2000 UV-Vis Spectrophotometer at 260 and 280 nm. The processed DNA was stored at -20 ℃.
The microbial diversity was analyzed by polymerase chain reaction (PCR) amplicons on the Illumina MiseqPE300 platform. The V3-V4 region of bacterial 16S rRNA genes were amplified by the primer pair 338F(5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R(5’-GGACTACHVGGGTWTCTAAT-3’), and all the PCR amplifications were carried out with the automatic thermal cycler. The amplified products were analyzed by gel electrophoresis in 2% agarose gels.
1.3.6 HTS on the Illumina MiseqPE300 platform
Samples with bright strip between 400 and 450 bp were chose for further analyses. These PCR products were mixed in equimolar amounts, followed by purification of the mixture with AxyPrep DNA Gel Extraction Kit. Sequencing libraries were produced using Sequencing libraries produced using TruSeq®DNA PCR-Free Sample Preparation Kit,according to the manufacturer’s instructions, and index codes were added. The qualification of the library was carried out using the Quanti Fluor™-ST fluorimeter and Bioanalyzer 2100 system. The library was sequenced finally by Majorbio Company on an Illumina MiseqPE300 platform that generated 250 bp paired end reads.
1.4 Data processing and analyzing
The antibacterial activity ofε-PL was measured in triplicate, and sequencing analysis of each sample three times in parallel. The differences between means were evaluated by theT-test analysis procedure using SPSS 17.0 software.
2 Results and Analyses
2.1 Isolation and identification of spoilage microbial strains
It has been known that cultural plating can be biased to some extent possibly due to competition between different bacteria in the culture media and the inability of culture media to cultivate dead cells and viable but non-cultivable cells which however can be identified by culture-independent method[19]. In this experiment, the typical growing colonies were selected from the PCA medium plate, and 34 purified strains of fish-derived microorganisms were isolated. After colony morphology observation and 16S rRNA identification, 13 strains ofAeromonassp., 3 strains ofShewanellasp.and 1 strain ofPseudomonassp. were obtained, which were listed in Table 1.Among the isolated strains, the number of isolatedPseudomonasstrains was relatively small. The possible reason was thatPseudomonasrequires higher nutritional conditions, and the PCA medium could not meet the requirements ofPseudomonas.According to the familiarity with the known NCBI strains and whether the strain is easy to be isolated and cultured,Aeromonassp. (F1),Pseudomonassp. (F6) andShewanellasp. (F21) which were isolated and identified were used as the bacterial strains to study the inhibitory activity ofε-PL.PseudomonasandAcinetobacterwere reported to be the predominant bacterial of fish[20-21].Aeromonaswere found to be associated with the spoil-age of iced wild rainbow trout and vacuum-packaged minced sturgeon[22], and some reports also showed thatPseudomonasandShewanellawere the prevalent populations in aquatic products under aerobic conditions[23], those were similar to our results.
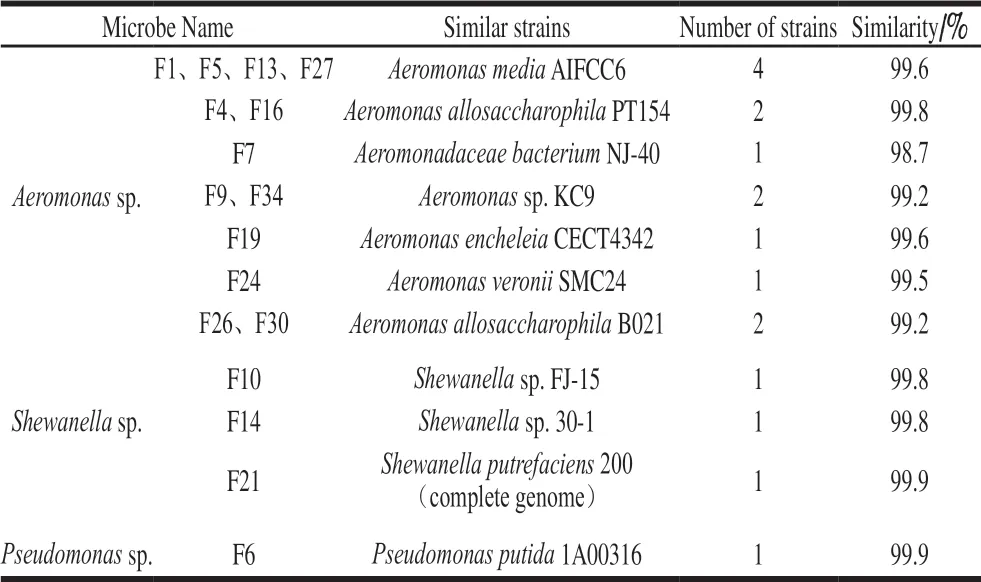
Table 1 Strains isolated from grass carp fillets
2.2 Antibacterial activity of ε-PL
The MIC ofε-PL onPseudomonassp.,Shewanellasp.andAeromonassp.were shown in Table 2. In the present study,ε-PL showed different antimicrobial activities against the tested strains based on the calculated MIC. The MICs forε-PL ranged from 0.50 to 1.00 mg/mL.Pseudomonassp. andShewanellasp. exhibited the least resistance againstε-PL compared withAeromonassp., showing a MIC of 0.5 mg/mL.MIC ofε-PL antibacterial activity againstAeromonassp.was found to be 1.0 mg/mL, which were higher than the MIC ofε-PL againstE. coliandListeria monocytogenesreported in the previous study[24-25]. Liu Wei found that the MIC ofε-PL againstPseudomonas aeruginosa(ATCC9027) andShewanella putrefaciens(ATCCBAA-1097) were 160 and 80 μg/mL respectively[26], which was lower than the results of this study. The reason may be that different sources of strains have different sensitivity toε-PL.
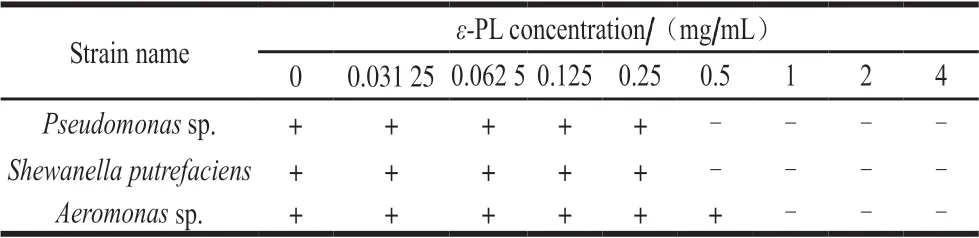
Table 2 MICs of ε-PL against Pseudomonas sp., Shewanella sp. and Aeromonas sp.
Changes in total viable counts andPseudomonascolonies of the fish fillets samples before or after being treated withε-PL were investigated (Fig. 1). According to the results of the study, there were no obvious difference in the total viable counts between groups treated with different concentration ofε-PL during the whole storage time. The treated samples had lower total colony counts compared with controlled group throughout the whole storage period. Especially on Day 7, the total colony counts inε-PL treated samples was around 5.5 (lg(CFU/g)) treated with 1.0 mg/mLε-PL, while the controlled samples reached 6.8 (lg(CFU/g)). In the same time (on Day 7) thePseudomonascolonies of treated(5.0 mg/mLε-PL) and untreated samples (CK) was 4.2 and 6.4 (lg(CFU/g)), respectively. Under the conditions of 1.0 mg/mLε-PL, the fresh grass carp fillets exceeded the secondary freshness limit (106CFU/g) on Day 9. The result showed that the shelf life of grass carp fillets treated with 1.0 mg/mLε-PL can be extended for 3-4 days according to the total viable counts, which was similar to the findings of Yu Dawei et al[27].
ε-PL was also documented as a natural and effective antibacterial agent for fish-spoilage bacteria. Nowadays,food-borne pathogens have been regarded as one of the major reasons for fish spoilage and a major threat to fisheries development[28].ε-PL is widely used for its superior performance of antimicrobial activity against many food-borne pathogens, includingEscherichia coliO157:H7,Staphylococcus aureusandBacillus subtilis[29-30]. As expected, most fish-spoilage bacteria were also suppressed byε-PL with its concentration of 1.0 mg/mL.

Fig. 1 Total viable counts of spoilage bacteria in grass carp fillets treated with ε-PL during (4 ± 1) ℃ storage
2.3 Sequencing analysis
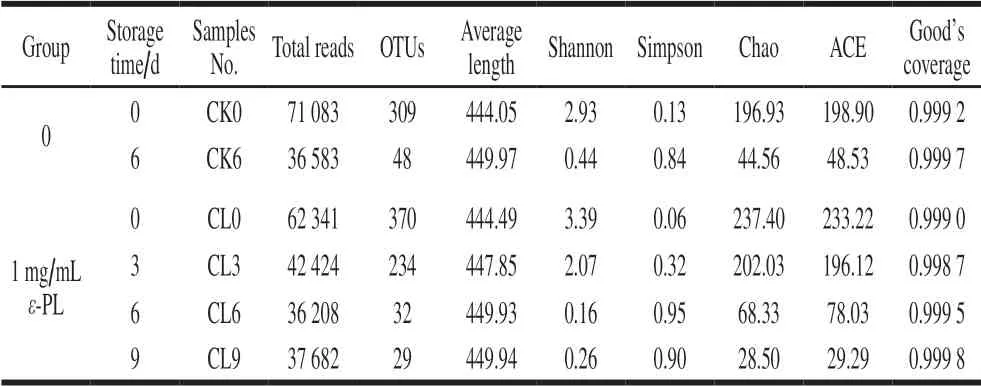
Table 3 Comparison of alpha diversity estimation of the 16S rRNA gene libraries by sequencing on an Illumina MiseqPE300 platform in modified atmosphere packaged grass carp fillets with and without 1 mg/mL ε-PL pretreatment during storage at 4 ℃
Valid sequences using the Illumina MiseqPE300 platform were obtained a total of 322 997 sequences originated from all controlled group and 535 967 sequences originated from treated group. The average value of those sequence was 447.01 and 448.05 reads/sample for controlled and treated groups, respectively. The length of generated sequence is mainly distributed in the range of 441-460 bp.The rarefaction analysis and the estimated sample coverage indicated that the amount of sequencing data was reasonable, and the amount of sequencing data was enough to represent the vast majority of microbial diversity information in samples. As presented in Table 3, Good’s coverage values for all the samples was higher than 99.9%, illustrating that there was satisfactory coverage for almost all the bacterial phylotypes in the fish samples.And the microbial species richness indexes such as ACE and Chao decreased with the extension of storage time. Indexes of Alpha diversity, including Sob, Shannon, Simpson, ACE,Chao and Coverage, were used to evaluate the richness and diversity of microbial species in fish samples according to the information of OTUs analysis[31].
2.4 Bacterial composition in grass carp fillets
2.4.1 Relative abundance at the phylum level
High-throughput sequencing based on the Illumina HiSeq300 platform quantified the composition and relative abundance of bacterial communities in grass carp fillets during refrigerated storage and the composition and relative abundance of bacterial communities were obtained by RDP classifier software. A total of 858 964 bacterial sequences and 388 different genera belonging to 28 phyla were observed and the relative abundances of the top 10 phyla in all samples at different times of storage are shown in Fig. 2. It shows that the predominant bacterial phyla in all the fish samples on Day 0, whether treated withε-PL or not, is consisted of Proteobacteria primarily accounting for 65.24% in CK group. Besides, a small amount of Actinobacteria (17.86%),Bacteroidetes (8.42%), Firmicutes (5.66%) and Cyanobacteria were also detected in CK group on Day 0.
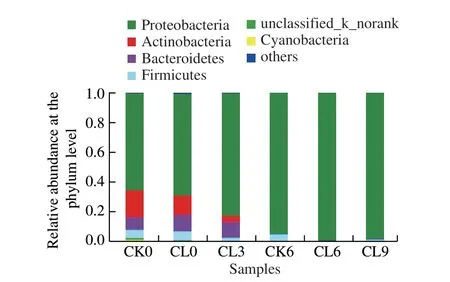
Fig. 2 Relative abundance at the phylum level based on the classification of partial 16S rRNA gene sequences of microbiota from grass carp fillets without and with 1.0 mg/mL ε-PL pretreatment during storage at 4 ℃
With the prolonging time of storage inε-PL treated group, the relative abundance of Proteobacteria increased gradually, while that of Actinobacteria, Bacteroidetes,Firmicutes and other bacterial phyla reduced. With treatmentε-PL for 9 days at 4 ℃ storage, the relative abundance of Proteobacteria increased from 68.45% to 99.20%. However,the relative abundances of Actinobacteria and other bacterial phyla were reduced from 12.93%, 6.38% to approximately zero, respectively. Meanwhile, relative abundances of Bacteroidetes and Firmicutes were found to decrease from 11.07%and 5.79% to 0.38% and 1.01%, respectively. To compare CK0 and CL0 samples, addingε-PL did not induce a significant change of bacteria communities except forCyanobacteria, which is only detected in CK0. After storage for 6 days, Firmicutes still could grow in CK6, but was inhibited byε-PL and disappeared in CL6.
2.4.2 Relative abundance at the genus level

Fig. 3 Relative abundance at the genus level based on the classification of partial 16S rRNA gene sequences of microbiota from grass carp fillets without and with 1.0 mg/mL ε-PL pretreatment during storage 4 ℃
A total of 388 different genera were observed and the relative abundances of major bacteria species at the genus level of microbiota from grass carp fillets without and with 1.0 mg/mLε-PL pre-treatment during 4 ℃ storage were presented in Fig. 3. Firstly, the bacteria diversity at the genus level of treated samples was found to be obviously decreased with storage time increased. In CK0,Pseudomonaswas the predominant bacterial (relative abundance was 21.45%),followed byAcinetobacter(10.31%) in the microbiota.Compared with the control group, the relative abundance ofPseudomonaswas found to decrease to 11.10% on Day 0,indicating that theε-PL pre-treatment could inhibitPseudomonasgrowth. Furthermore,Pseudomonasincreased rapidly under refrigerated storage both treated group and controlled group with storage time increased, and became the predominant bacterial genus (91.55% in controlled group on Day 6 and 95.04% in treated group on Day 9, respectively).

2.5 Effect of ε-PL pre-treatment on microbial diversity of grass carp fillets
Fig. 4 shows the effects ofε-PL pre-treatment on the initial microbial community diversity at the genus level. The result showed that the relative abundance ofPseudomonasdecreased to varying degrees withε-PL pre-treatment (from 21.46% to 11.0%),Chryseobacterium,Glutamicibacter,Kocuria,Pantoea, etc.. Whereas, the relative abundances ofAcinetobacter(from 10.31% to 12.46%),Ralstonia,Burkholderia-Paraburkholderia,Psychrobacter, etc. were increased. Results indicated thatε-PL had different inhibition ability on various species of microorganisms in real food matrix.
After 6 days storage at 4 ℃, microbial community changed dramatically (Fig. 5). The relative abundance ofPseudomonasreached 97.47% inε-PL pre-treatment group,and other five types of microtia had the relative content was more than 0.1%, includingJanthinobacterium(0.64%),Chryseobacterium(0.56%),Shewanella(0.44%),Acinetobacter(0.31%),Comamonadaceae(0.15%). Nevertheless,Pseudomonaswas also the major microbial with the relative abundance value of 91.55% in the controlled groups on the 6th day at 4 ℃ storage. Besides,Pseudomonas,Brochothrix(2.96%),Acinetobacter(1.97%), andPsychrobacter(0.20%) etc.were also detected. Those results showed thatε-PL pre-treatment effectively inhibited the growth ofAcinetobacter,Brochothrix,Shewanella, andPsychrobacter.
T-test analysis was used to analyze the significance change of microbial community afterε-PL pre-treatment (Fig. 4C and Fig. 5C). Results showed thatε-PL pre-treatment had extremely significant influences onFlavobacteriumin fresh grass carp fillets(P< 0.01), and significant influences onPseudomonasin grass carp fillets after storage 6 days (P< 0.05).
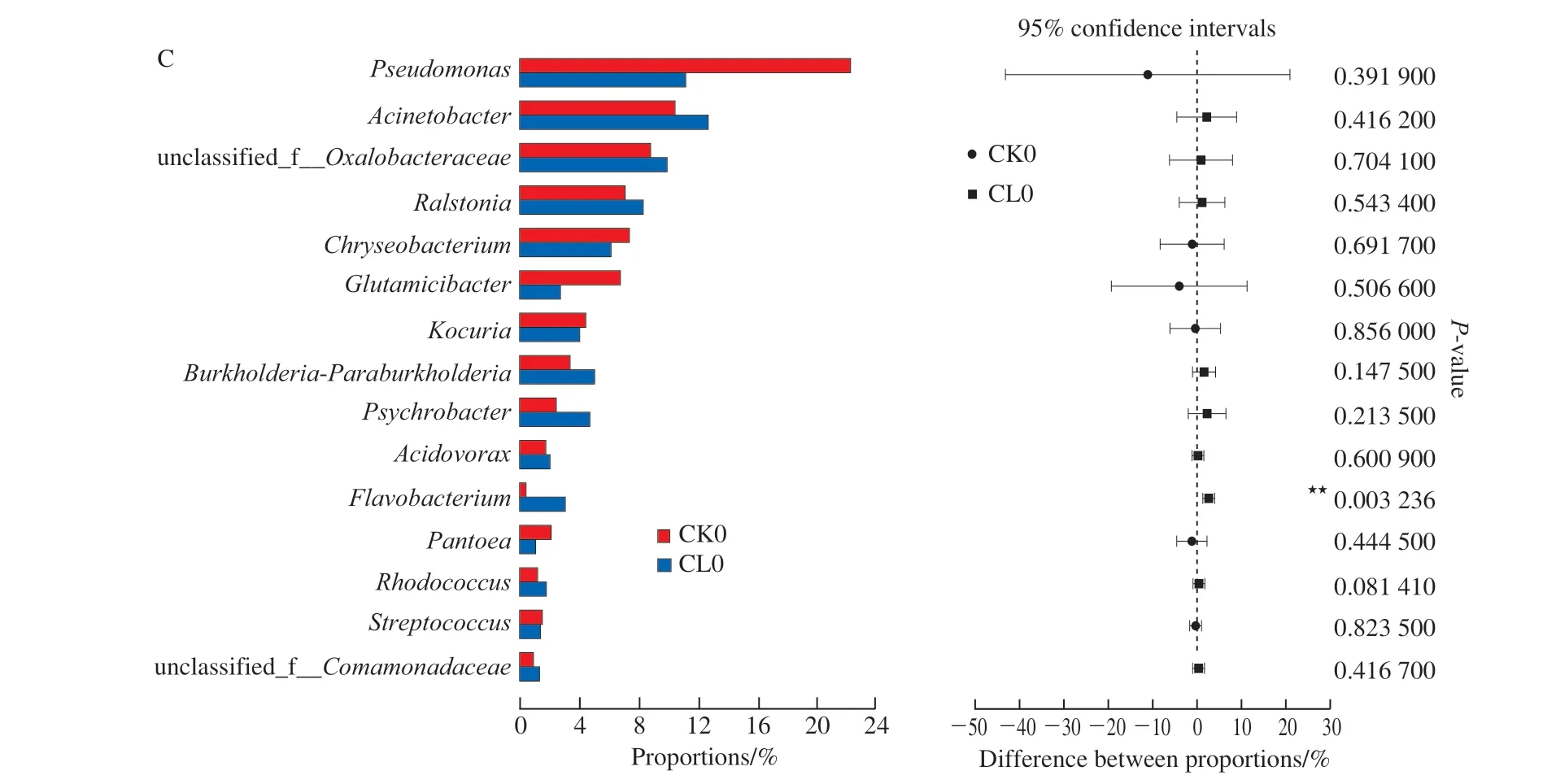
Fig. 4 Initial microbial community diversity at the genus level in fresh grass carp fillets from control (A) and ε-PL pretreatment groups (B),and T-test analysis (C) between them
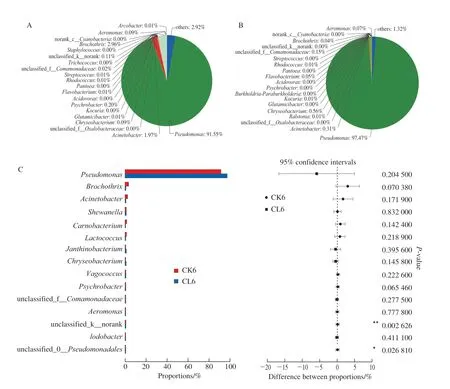
Fig. 5 Microbial community diversity at the genus level in 6-day-stored grass carp fillets from control (A) and ε-PL pretreatment groups (B),and T-test analysis (C) between them
3 Conclusions
The growth ofPseudomonassp.,Shewanellasp. andAeromonassp. which were isolated and identified from grass carp fillets, were inhibited byε-PL with the concentration of 1.0 mg/mL. The microorganism species in the sample decreased continuously, especially after being stored for 6-9 days, and the microbial diversity of grass carp fillets withε-PL pre-treatment was decreased with the storage time increased. The relative abundance ofPseudomonasincreased rapidly under refrigerated storage whether treated group or controlled group, andPseudomonasbecame dominant in the spoilage progress of grass carp fillets, which was the major contributor to grass carp fillets decomposition. The HST technology was proved to be a valuable and promising tool on assessing the effects of external treatments on microbial diversity in fish samples, which will be helpful in understanding the relative spoilage dynamics in food and provide theoretical basis for quality and safety monitoring, as well as oriented control of the spoilage bacteria.
