Regeneration Function Analysis of GmESR1 in Transgenic Soybean
2020-07-15LiMoPanXiaochengLiuYangLiYuanmingLiHongweiLiuWeiSuAnyuandWuXiaoxia
Li Mo, Pan Xiao-cheng, Liu Yang, Li Yuan-ming, Li Hong-wei, Liu Wei, Su An-yu, and Wu Xiao-xia, *
1 College of Agriculture, Northeast Agricultural University, Harbin 150030, China
2 65301 Farm and Sideline Bases of Army, Wudalianchi 164100, Heilongjiang, China
3 College of Resources and Environmental Sciences, Northeast Agricultural University, Harbin 150030, China
4 College of Heilongjiang Green Food Research Institute, Harbin 150030, China
Abstract: ENHANCER OF SHOOT REGENERATION (ESR1) is an important regulator of plant regeneration in vitro, which promotes regeneration of plant. In this study, transgenic positive plants with normal expression of proteins were screened by molecular assay. Through the study of the transgenic plants and the control Dongnong 50, the difference between immature embryo-induced callus and induced shoot bud was observed. The increase in callus weight indicated that GmESR1 gene accelerated the formations of shoot buds. By measuring the changes of hormone in the process of induction callus of transgenic plants, it was found that the contents of indole-3-acetic acid (IAA) and zeatin (ZT) in transgenic lines were significantly increased. It could be concluded that GmESR1 gene promoted the accumulation of hormone and affected regeneration process. In addition, this study also veri fied the interaction between GmBIM1 gene and GmESR1 gene by bimolecular fl uorescence complementation (BiFC).
Key words: transgenic soybean, GmESR1, regeneration, indole-3-acetic acid (IAA), zeatin (ZT)
Introduction
ESR1 (ENHANCER OF SHOOT REGENERATION;DORNRÖSCHEN) gene belongs to the AP2/EREBP(APETALA2/ethylene-responsive element binding protein) family (Bannoet al., 2001). The gene of reactions participates in various biological functions,throughout the growth and development of plants,and also makes respond to adverse environmental signals, such as high salinity and drought. This family of transcription factors is plant-specific family and has not less than 144 family members (Sakumaet al.,2002).ESR1 encodes a transcription factor belonging to the ERF family of the EREBP subfamily. The ERF family is the largest branch of the AP2/ERF(APETALA2/ethylene-responsive factor) family and contains one or two AP2/ERF domains with specific DNA binding motifs (Nakanoet al., 2006; Shigyoet al., 2006). The region, which has the AP2/ERF domain and the ESR-specific motif ofESRgene, is considered to be a necessary part in enhancing shoot regeneration (Nomuraet al., 2009).ESR1 acts as a transcriptional activating substance (Matsuoet al.,2011; Matsuo and Banno, 2008). At the same time, the ERF domain ofESR1 gene, which is closely related to the induction of cytokinin, speci fically binds to thecisacting element of the GCC box (GCCGCC) encoding the ethylene response system, so it is considered that the transcribed region ofESR1 gene involves in response to ethylene signaling pathway (Zhanget al.,2017).
Bannoet al. (2001) showed that in the presence of exogenous cytokinins, overexpression ofESR1 can greatly improve the efficiency of root explant regeneration.ESR1 gene promotes the induction of callus duringin vitroregeneration.ESR1 is activated byWIND1 (WOUND INDUCED DEDIFFERENTIATION)gene (Okamuroet al., 1997; Delessertet al., 2004),mediates wound-induced callus formation by grade transcription and promotes subsequent shoot regeneration (Iwaseet al., 2016). Under the condition of plant injury,WIND1-4 promotes the formation of callus at the wound site by activating cytokinin signaling(Iwaseet al., 2011(A); Iwaseet al., 2011(B)); in the development of tissues,ESR1 affects the homeostasis of stem cells in meristematic tissues by participating in the two pathways. The first pathway, relying onSTM(SHOOTMERISTEMLESS) gene regulation(Spinelliet al., 2011; Hakeet al., 2004; Hamant and Pautot V, 2010; Hay and Tsiantis, 2010), which promotes cell division, primarily controlling cells in an undifferentiated state and inhibiting differentiation(Lenhardet al., 2002; Endrizziet al., 1996; Longet al.,1996).STM, one of the potential target genes, is regulated byESR1. According to Kirchet al. (2003),ESR1 can inhibit the expression ofSTM, promote cell differentiation and meristem development. The second pathway is a feedback regulation loop formed by CLV-WU (CLAVATA-WUSCHEL), which can maintain dynamic balance of stem cells in shoot tip meristems (Mayeret al., 1999; Schoofet al., 2000). Its mechanism of action in plants is thatCLVgene (Brandet al., 2000; Fletcheret al., 1999) affects the size of the stem cell microenvironment in the stemend meristem by inhibiting the expression ofWUSgene (Lauxet al.,1996; Mayeret al., 1999). Kirchet al. (2003) showed thatESR1 gene was involved in the CLV-WUS feedback regulation loop, which in turn affects the development of meristematic central stem cells. In addition, previous studies have also found thatESR1 affects development of embryonic shape by interacting withBIM1 (BES interacting Myc-like protein 1) gene inArabidopsis(Chandleret al., 2009).
In these processes, cytokinin and auxin play a vital role. In the development of meristem, auxin and cytokinin synergistically regulate stem cell homeostasis(Luoet al., 2018). At the same time, hormone is important forESR1invitroculture condition (Coleet al., 2009; Fanet al., 2012). OverexpressingESR1 can produce shoots by usingArabidopsis thalianaroot explants under the presence or absence of cytokinin,but in the case of a large number of cytokinin, the efficiency of shoot regeneration is significantly improved (Bannoet al., 2001).
Materials and Methods
Molecular detection of transgenic plants
Specific primers were used in PCR amplification ofBargene sequences, which used transgenic line DNA as a template (Bar-F: 5'-ATATCCGAGCGCCTCGTG CAT-3'; Bar-R:5'-GGTCTGCACCATCGTCAACCA CT-3'); the procedure was as the followings: 95℃,3 min; 95℃, 15 s; 58℃, 30 s; 72℃, 30 s, 35 cycles,final extension 72℃, 7 min, 4℃, cooling (Wanget al.,2018).
TRIzol reagent was used to extract the total RNA from transgenic lines and the control Dongnong 50(DN50); and cDNA was synthetized by qRT-PCR reagents including HiscriptII Q RT Super Mix and qPCR+gDNA wiper kit (all the experimental steps followed the instructions). Reaction condition was:95℃ for 5 min; 95℃ for 10 s, 62℃ for 30 s, 72℃ for 30 s, 40 cycles; dissolution curve, 95℃ for 5 s, 65℃for 1 min, 97℃ for 30 s; 4℃, cooling. TheCTvalue was calculated by the 2-ΔΔCTmethod and three technical replicates were performed for each sample. Specific primers were RTGmESR1-F: 5'-CTTCCACTCAGAA CTTCCACGAC-3'; RTGmESR1-R: 5'-TAACAGACA AAGAGCCTCCACAA-3';GmActin-F: 5'-GTGTCAG CCATACTGTCCCCATTT-3';GmActin-R: 5'-GTTT CAAGCTCTTGCTCGTAATCA-3'. The difference in the expression level ofGmESR1 gene among different tissues of the transgenic lines was determined.
Analysis of transgenic regeneration function
The callus was induced by 5 mm immature embryo from the transgenic and the control lines in the field.Each transgenic line was cultured five bottles and six immature embryos per flask were cultured. Callus induction rate was counted after 20 days. After the calluses were induced, the calluses of each line were excised and transferred to the subculture medium for futher culture. Twenty bottles were cultured per line and all of the calluses were transferred to subculture for 40 days. Calculated the callus weight for each line.The auxin (IAA) and cytokinin (ZT) were measured by ELISA kit.
Using soybean cotyledon node as explants cultured soybean plants. The difference between the transgene and the control plants was observed. Counted the weight. Another part of explants was cut with a scalpel to remove the cotyledon node. Only the buds were gently placed into the elongation medium with tweezers and the shoot elongation induction culture was carried out, when the buds were elongated to about the mouth of the bottle. Compared the number of shoot elongation between the transgene and the control, according to the count data.
GmESR1 gene and GmBIM1 gene interaction veri fication
Full-length PCR amplification ofGmESR1 gene andGmBIM1 gene were carried out and the amplified product was ligated into the pGWC vector (entry vector contained attL1 and attL2). The target gene was ligated into pEarlyGate201 and pEarlyGate202 by LR reaction, respectively. The plasmid was transformed intoE. coliDH5αand sequenced. The correctly sequenced bacterial was extracted plasmid, which needed to be transformed into EHA105. Specific primers wereGmESR1-F 5'-ATGAGGCGTCTCAACG GGGT-3';GmESR1-R 5'-CCAGCATTTTGCATTCTGA T-3';GmBIM1-F 5'-ATGGAGCTTCCTCAAGCACG-3';GmBIM1-R 5'-CCACTTCTGCAAGTCTTTAG-3'.
The EHA105 was injected into the growing tobacco and the fl uorescence expression was observed by confocal microscopy after 3 days (Bhatet al., 2006).
Results
Molecular identi fication of transgenic soybean
The results of PCR amplification of the transgenic lines and the control DN50 plant are shown in Fig. 1A.Five positive plants were obtained by the speci ficBargene. Subsequently, qRT-PCR analysis of the five positive plants showed that the relative expression level ofGmESR1 gene in the four plants was higher than that of the control DN50 (Fig. 1C). To further test whether the transgenic gene could normally express the protein, the transgenic plants were subjected to Western blot hybridization experiments (primary antibody was Bar antibody). The results showed that these four transgenic lines were able to express Bar protein normally (Fig. 1D). The result indicated thatGmESR1 gene was stably inherited and normally expressed.
Analysis of transgenic regeneration function
The immature embryos of the transgenic soybean lines and the control DN50 were as explants and the soybean callus was obtained in the induction medium (Fig. 2A).The induction rate was investigated after 20 days and the callus was transferred to subculture medium.After induction for 40 days, the weight of callus was counted and the test results were significantly different. The results are shown in Table 1. After induction of young embryos for 20 days, all the callus induction rates of the transgenic lines were higher than those of the control DN50. After 40 days of callus subculture, the callus weight of the transgenic plants was significantly higher than that of the control DN50.The qRT-PCR analysis of different transgenic tissues revealed that the expression ofGmESR1 gene was the highest in immature embryos than in other tissues(Fig. 2B). It was speculated thatGmESR1 gene was important for the subsequent development of immature embryos. At the same time, the total RNA of immature embryos and calluses from DN50 and the transgenic soybean were extracted. The relative expression ofGmESR1 gene was analyzed by qRT-PCR (Fig. 2C),according to the results, the relative expression level ofGmESR1 gene in calluses was higher than that in young embryos. It indicated that immature embryos of the transgenic lines were superior to that of the control DN50 in the induction of callus provliferation.

Fig. 1 Molecular detection of transgenic plants

Fig. 2 GmESR1 expression analysis and callus induction analysis of transgenic soybean immature embryos and callus
Using transgenic soybean and the control DN50 cotyledon node as explants to culture shoots, the cluster buds were cultured and weight was statistically analyzed. It was found that the cluster buds of the four transgenic lines were larger than those of the control DN50 (Fig. 3A). The weight was significantly higher than that of the control DN50 (Table 2); the number of regenerated seedlings by statistical cluster bud elongation (Fig. 3B, Table 2) indicated that the transgenic soybean increased the number of cluster buds, thereby increasing the regenerated seedlings.
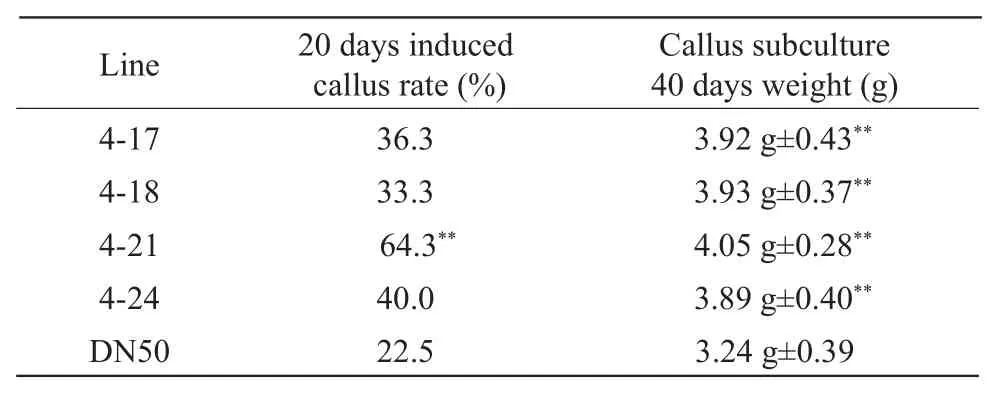
Table 1 Analysis of transgenic GmESR1 soybean callus induced and gravimetric of callus proliferation

Fig. 3 Phenotypic analysis of transgenic plants
Changes of hormone contents in callus induced by transgenic GmESR1 gene
To understand the changes of hormone in the process of inducing callus, IAA and ZT in young embryos and callus of the transgenic plants and the control DN50 were determined by ELISA. In Fig. 4A, the content of IAA was increased, during the induction of callus;and the content of IAA in the immature embryos and callus of the transgenic lines were higher than those in the control DN50. In Fig. 4B, the content of ZT of the transgenic lines was increased in the process of immature embryo-induced callus; while ZT content in the control DN50 decreased slightly, during the induction process. The above results indicated thatGmESR1 might lead to the accumulation of IAA and ZT, during callus induction, thereby promoting the proliferation of callus and enhancing the regeneration ability of transgenic lines.

Table 2 Analysis of transgenic GmESR1 soybean weight of cluster shoots and number of induced seedlings
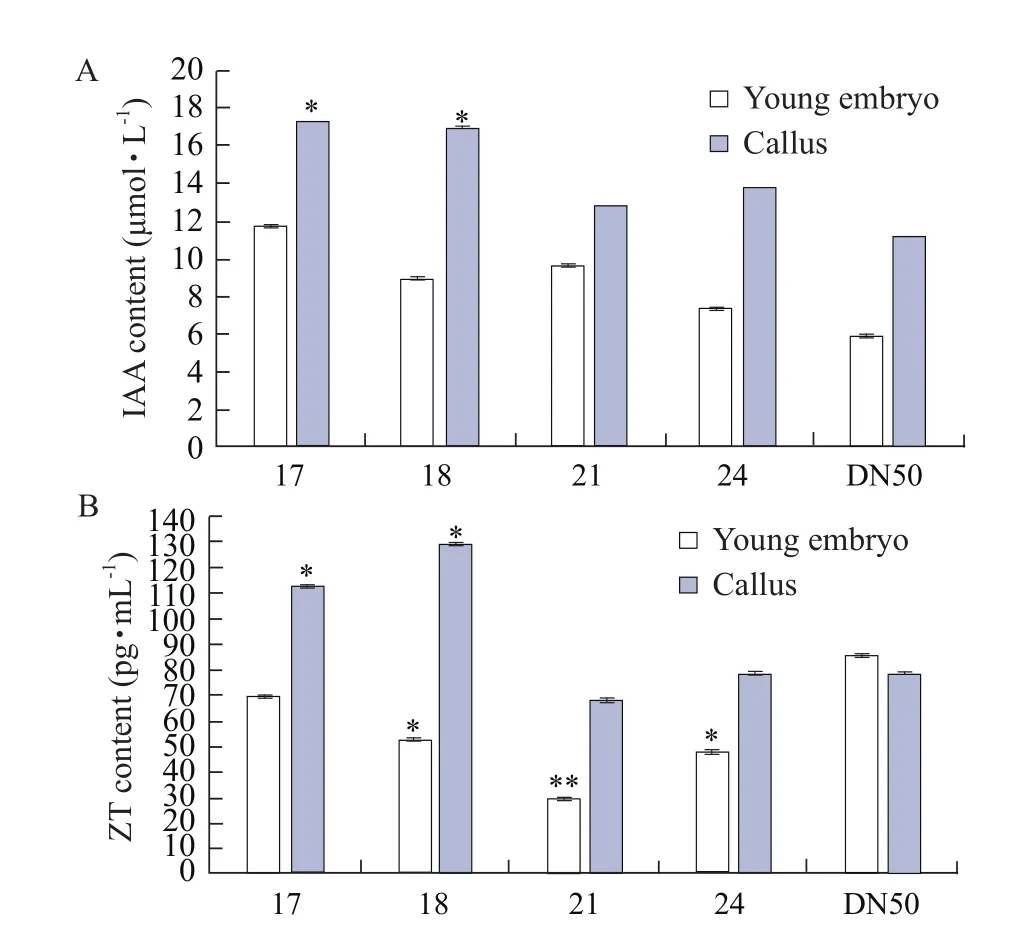
Fig. 4 Hormone content in transgenic immature embryos and callus
Binary molecular fluorescence complemention analysis of GmESR1 and GmBIM1 interaction
According to the laboratory's preliminary screening of interaction genes by pre-laboratory yeast twohybrid experiment, a proteinGmBIM1 interacting withGmESR1 protein was found. To further verify whetherGmESR1 andGmBIM1 proteins could interact in plant cells,GmESR1 andGmBIM1 were constructed into plant expression vectors pEarlyGate201 and pEarlyGate202, respectively. The recombinant plasmid was successfully constructed by bacterial PCR and sequencing (Fig. 5A).

Fig. 5 BiFC analyses of GmESR1 interaction with GmBIM1
Agrobacteriumcarrying the recombinant plasmid was injected into tobacco and the yellow fluorescent signal (YFP) in the epidermal cells of tobacco leaves was detected by fluorescence confocal microscopy.The results are shown in Fig. 5B, which indicated that a yellow fluorescent signal could be detected in epidermal cells in tobacco leaves co-transformed with pEarlyGate201-GmESR1 and pEarlyGate202-GmBIM1.
No yellow fluorescent signal could be detected by co-transformation of pEarlyGate201-GmESR1 and pEarlyGate202, pEarlyGate201 and pEarlyGate202-GmBIMI1. The results indicated thatGmESR1 andGmBIMI1 proteins could be interacted in plant cells and interact with each other in the cell membrane.
Discussion
This study further cleared the role ofGmESR1 gene in the regeneration process by investigating the difference between induction of callus and cotyledon node to shoot by using the transgenic lines and control DN50 plants. According to previous studies, it was known that the regulator which rapidly activated and played a key role in the process of inducing callus formation in the wound was a subfamily ofWOUND INDUCED DEDIFFERENTIATIONin theAP2/ERFtranscription factor (WIND1,WIND2,WIND3 andWIND4). By constructingWIND1-SRDX(deficientWINDfunction)mutants, the researchers found that the mutant plants strongly inhibited and blocked adventitious bud regeneration (Iwaseet al., 2015), andESR1 gene inWIND1-SRDXplants could compensate for this defect. The study found thatWIND1 directly bound to theESR1 promoter and activated its expression,promoting callus formation and regeneration of shoots to cope with wound effect (Iwaseet al., 2016). This study investigated the weight and induction rate and found thatGmESR1 gene promoted the formation of callus.
According to Iwaseet al. (2016), auxin and cytokinin acted withESR1 gene duringin vitroregeneration,synergistically. However, the callus of the transgenic lines showed accumulation of hormones. It was concluded thatGmESR1 gene was not only regulated by hormonal signals, but also regulated the hormone signal, more effectively promoted gene expression and improved soybean regeneration. According to BiFC and yeast two-hybrid experiments betweenGmBIM1 andGmESR1, although experiments did not verify the significance of interaction, previous studies found that the interaction ofESR1 andESR2 withBIM1 affected embryonic development, providing environmental conditions for the subsequent embryo growth (Chandleret al., 2009).
Based on the research results of this experiment,the next step would be to use the transgenic lines as materials to study the pathway ofGmESR1 genein vitroregeneration and the path of action with hormones. Combining the work of this research with modern agriculture will be conducive to the application of genetic modification in agriculture, it will alleviate the problem of low transgenic efficiency of soybean and accelerate the application of genetic modification in agriculture.
Conclusions
The specific function of the soybeanGmESR1 gene was analyzed and the formation of callus and adventitious bud were promoted. In soybean, overexpression ofGmESR1 caused accumulation of IAA and ZT contents and promoted regeneration. These results indicated thatGmESR1-based soybeans were regeneratedin vitro.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Effect of Salt Stress on Nitrogen Assimilation of Functional Leaves and Root System of Rice in Cold Region
- Effects of Straw Returning with Different Tillage Patterns on Corn Yield and Nitrogen Utilization
- Effects of Rare Earth Lanthanum and Cerium on Antioxidant Enzyme Activities in Soybean Leaves
- Regulating Effect of Exogenous Silicon on Soil Fertility in Paddy Fields
- Uptake of B, Co and Ni by Plants from Oil Contaminated Soil Capped with Peat
- Anti-tumor Effect of Newcastle Disease Virus Expressing IL-2 in Lung Cancer Model
