Establishment and evaluation of a TaqMan RT-qPCR method for tissue quantification of oncolytic virus M 1 in vivo*
2020-06-09QINZhenDANJiaHUJunLIANGJiankaiYANGuangmeiCaiJing
QINZhen,DANJia,HUJun,LIANGJian-kai,YANGuang-mei,Cai Jing
(Department of Pharmacology,Zhongshan School of Medicine,Sun Yat-sen University,Guangzhou 510080,China.E-mail:caij53@mail.sysu.edu.cn)
[ABSTRACT] AIM:To establish a TaqMan RT-qPCR method for surveiling the spread of oncolytic virus M1 in tissue,helping control the dosage and assessing the safety of virus.METHODS:A TaqMan-based one-step RT-qPCR method for the detection and quantification of oncolytic virus M1 in the tissues was established.The virus load and distribution in the tissues of SD rats,cynomolgus monkeys and nude mice were also investigated.RESULTS:A pair of specific primers(Q3)and the standard viral RNA for SYBR Green RT-qPCR were screened and selected with the best specificity and amplification efficiency.By optimizing the experiment conditions,we found that the annealing temperature above 62℃reduced matrix effect but affected the amplification efficiency.So we established a one-step TaqMan RT-qPCR method and redesigned a pair of Q3 short primers(Q3S).Using the one-step TaqMan RT-qPCRand Q3Sprimer,the standard RNA with low copy numbers was specifically detected under the background of mixed matrix RNA of SD rats or cynomolgus monkeys.Furthermore,the method was verified to be suitable for detecting tissue distribution of M1 virus in the mice,SD rats and cynomolgus monkeys.CONCLUSION:The TaqMan-based one-step RT-qPCR constructed with Q3S primer can be used for M1 virus quantification in various tissue samples of different animals with better specificity and sensitivity,and may be further applied to the detection of clinical samples.
[KEY WORDS] TaqMan;RT-qPCR;Oncolytic virus M1
Despite the progress in tumor treatment in the past decades,cancer remains a major health problem in the world[1].Oncolytic virotherapy is a kind of innovative anticancer strategy which is based on natural or genetically engineered viruses specifically replicating in malignant cells rather than normal cells[2].Tumor's selectivity of oncolytic virus is mainly due to genetic abnormalities of malignant cells,such as innate immune defects and abnormally activated oncogenes[3-4].Oncolytic viruses lead to direct cell lysis,bystander killing and indirect anticancer immune response.Some of the oncolytic viruses have made rapid progress in pre-clinical and clinical trials[5].In 2015,the U.S.Food and Drug Administration(FDA)and the European Medicines Agency have approved talimogene laherparepvec(also known as T-Vec),which is derived from herpes simplex virus(HSV)and genetically engineered to express granulocyte-macrophage colony-stimulating factor(GMCSF)for the treatment of melanoma patients,making it the first and only oncolytic virus approved by the FDA for clinical therapy[6-7].
Previously we have identified M1 virus,a strain of alphavirus which was isolated from culicine mosquitoes in Hainan,China,as a novel oncolytic virus that selectively replicates in ZAP-defective human cancers and induces irreversible endoplasmic reticulum (ER)stress-mediated apoptosis[8-10].Importantly,M1 was reported to be non-pathogenic for nonhuman primates after multiple rounds of repeated intravenous injections[11].Moreover,we further found that some small molecular drugs dramatically enhanced the oncolytic effect of M1,including second mitochondria-derived acti-vator of caspase(Smac)mimetic compounds(SMCs),Bcl-xL inhibitors,and inhibitors of valosin-containing protein(VCP)[12-14].Therefore,it is indicated that M1 virus is a promising therapeutic candidate for cancer treatment.
Accurately quantifying the copy number of M1 virus in the samples of different tissues is of great importance to evaluate the efficiency and the safety of M1 virus in the clinical application.The traditional method of M1 viral RNA quantification is SYBR Green-based quantitative reverse transcription PCR(RT-qPCR),but its specificity and sensitivity are not enough.Therefore,a more sensitive and specific quantitative method need to be established urgently.In this report,we developed a TaqMan-based RT-qPCR method for the quantification of M1 viral RNA in tissues.
MATERIALSAND METHODS
1 Ethics statement
Animal study was performed and approved by the Institutional Animal Care and Use Committee(IACUC)at JOINN Laboratories,China.All experiments were conducted according to the U.S.“Public Health Service Policy on Humane Care and Use of Laboratory Animals”.
2 Virus and cell lines
The M1 virus of c6v1 strain used in this study was obtained from Vero cells and provided by Guangzhou Virotech Pharmaceutical Technology Co.LTD.The virus titer was determined by TCID50 assay using BHK-21 cells.BHK-21 cells and Hep3B cells were from American Type Culture Collection(ATCC),and were passaged according to the guidelinesof ATCC.
3 Experimental procedures
3.1 ViralRNAextractionViral RNA was extracted from thawed virus lyophilized powder samples using GeneJET RNA Purification Kit(Thermo Fisher Scientific)according to the manufacturer's instructions.The extracted viral RNA was stored at-70℃.
3.2 MatrixRNAextractionThe matrix RNAs were extracted from different sources of tissues,including serum,excrement,urine and liver.The matrix RNAs from cynomolgus monkey and rat's tissues were extract using GeneJET RNA Purification Kit according to the manufacturer's instructions.The concentration of extracted matrix RNA was diluted to 0.5 g/L and was stored at-70℃.Before using frozen RNA samples,absorbance at 260 nm was determined,and the concentration of total RNA was calculated.The recovery rate(%)=the mean amount of thawed RNA/theoretical amount of the measured RNA×100%.The extracted matrix RNA was used when the recovery rate was between 90%~110%.Otherwise,the RNA should be reextracted.
3.3 Design and synthesisofSYBR Green primersThree M1-specific SYBR Green primers were designed in Beacon Designer.Table 1 showed the primer sequences.All primers were compared among human,cynomolgus monkey and rat in primer BLAST for specificity analysis.

Table 1.The primer for detecting M 1 virus by one-step SYBR Green RT-qPCR method
3.4 Design and synthesis of standard viral RNAsThe instability of RNA in M1 viral particle makes it unqualified as standard RNA for RT-qPCR,hence synthesized RNA was used to replace it as the standard RNA.According to SYBR Green primers,we designed the standard viral RNA Q1 and Q3.The standard RNA was synthesizedinvitroby GenScript.Table 2 showed the standard RNA sequences.
3.5 Design and synthesis of TaqMan primersAccording to standard RNA Q3,the TaqMan primers and one TaqMan probe were designed,and synthesized by GenScript.Table 3 showed the sequences of the primers and probe.
3.6 One-step SYBR Green RT-qPCRThe onestep SYBR Green RT-qPCR was performed with One Step TB Green™ PrimeScript™ PLUSRT-PCR Kit(Ta-KaRa)according to the manufacturer's instructions.The total reaction volume was 20.0μL,consisting of1× One Step TB Green RT-PCR buffer,PrimeScript PLUS RTase Mix,TaKaRa Ex Taq HS Mix,forward and reverse primers(0.4 μmol/L),viral standard RNA and matrix RNA.All amplification was performed in duplicate using the CFX96™Real-Time PCR Detection Systems(Bio-Rad)with the following amplification cycles:reverse transcription at 42℃ for 5 min,initial denaturation at 95℃ for 10 s,45 cycles of 95℃ for 5 s and 60℃ or 62℃ for 20 s,and melting curve analysis from 65℃ to 95℃.Raw data was analyzed to determine the amount of viral RNA based on the cycle threshold(Ct)values.The efficiency of RT-qPCR was measured fromthe slope of the standard curve.

Table 2.M 1 virus standard viral RNAs

Table 3.The primers and probes for detecting M 1 virusby one-step TaqMan method
3.7 One-step TaqMan RT-qPCRThe one-step TaqMan RT-qPCR was performed with FastKing One Step RT-qPCR Kit(TIANGEN)according to the manufacturer's instructions.The total reaction volume was 25.0 μL,consisting of 1× FastKing One Step Probe RT-qPCR Master Mix,1× FastKing Enzyme Mix,forward and reverse primers(0.4 μmol/L),TaqMan probe,viral standard RNA and matrix RNA.All amplification was performed in duplicate using the CFX96™Real-Time PCR Detection Systems(Bio-Rad)with the following amplification cycles:reverse transcription at 50℃ for 30 min,initial denaturation at 95℃ for 3 min,and 45 cycles of 95℃ for 15 s and 60℃ for 30 s.Raw data was analyzed to determine the amount of viral RNA based on the cycle quantification(Cq)values.The efficiency of RT-qPCR was measured from the slope of the standard curve.
3.8 Tissue distribution of M 1 virus in ratsSD rats of 6~7 weeks old were from JOINN Laboratories and randomly grouped.The SD rats in the administra-tion group were intravenously injected with 2.28×107CCID50/animal(n=10+5)of M1 virus(c6v1 strain).The administration volume was 0.1 mL/animal.The administration speed was 1~2 mL/min.Three rounds of administration were performed.Each round included 5 d of treatment(i.v.,qd),and each interval was 14 d.After the 15th injection,the rats were raised to recover for 4 weeks.All rats were weighed before grouping(day-1)and continued weighing during the administration period on days 5,12,19,24,31,38 and 43.Subsequently,they were weighed once a week during the recovery period.The temperature was measured before grouping(day-1)and continually on days 1,5,20,43 and 71.The total RNAs of different tissues were extracted on day 44(n=10)and day 72(n=5)by viral RNA extraction kit(Promega).Viral copy numbers were determined by TaqMan RT-qPCR with Q3S primer.In addition,the pathological changes were examined on day 72(n=5).
3.9 Tissue distribution of M 1 virus in monkeysThe cynomolgus monkeys(2~5 years old) were obtained from JOINN Laboratories and randomly grouped.The cynomolgus monkeys in the administration group were intravenously injected with 3.29×107CCID50/animal(n=3+3)of M1 virus(c6v1 strain).The administration volume was 0.5 mL/animal.The administration speed was 2.5~3 mL/min.The M1 virus administration scheme was similar to that of SD rats,i.e.administration on days 1~5,days 20~24 and days 39~43 once a day for a total of 15 times.All monkeys were weighed before grouping(day-3)and continued weighing during the administration period on days 5,12,19,24,31,38 and 43.Subsequently,they were weighed once a week during the recovery period.The temperature was measured before grouping(day-3)and continually on days 1,5,20,43 and 71.The total RNAs of different tissues were extracted on day 44(n=3)and day 72(n=3)by viral RNA extraction kit(Promega).Viral copy numbers were determined by Taq-Man RT-qPCR with Q3Sprimer.In addition,the pathological changes were examined on day 72(n=3).
3.10 Tissue distribution of M 1 virus in nude miceThe BALB/c nude mice of 8~9 weeks old were obtained from JOINN Laboratories.The Hep3B cells(5×106/mouse)were subcutaneously inoculated into the right armpit of the mice.When the tumor grew to around 100 mm3,the mice were randomly divided into 8 groups(n=6).The M1 virus(c6v1 strain)at dose of 6.75×109CCID50/kg was intravenously injected into the BALB/c nude mice.The administration volume was 25 mL/kg.The total RNAs were extracted from different tissues at 2,12,24,48,72,168,336 and 672 h after the injection of M1 virus by viral RNA extraction kit(Promega).Viral copy numbers were determined by TaqMan RT-qPCRwith Q3Sprimer.
3.11 Pathological examinationThe animals were euthanized at the indicated time point,and then different tissues were subjected to histological treatments,including paraffin embedding, sectioning, and HE staining.Diagnosis and classification of pathological changes were accessed using standard terminology by pathology experts from JOINN Laboratories.The degree of pathological changes under the microscope was divided into 4 levels:normal,mild,moderate,and severe.
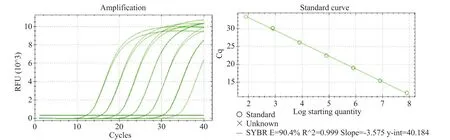
Figure 1.The specificity and amplification efficiency of Q3 primer for standard viral RNA detection.Gradient diluted standard viral RNA was detected by one-step SYBR Green RT-qPCR.The specificity and amplification efficiency of Q3 primers were showed.
4 Statistical analysis
All statistical analyses were performed using GraphPad Prism software.Comparisons between different groups were made using Student'sttest(2 groups)or analysis of variance(more than 2 groups)followed by Tukey's multiple comparisons.Data were represented as mean±SD of at least 3 biological repeat experiments.Pvalues of<0.05 were considered statistically significant.
RESULTS
1 Standard viral RNAsand primer design
It's known that the maximum length of synthetic RNA is ~100 nt,so we designed 3 pairs of primers(Q1,Q2 and Q3)whose amplification product lengths were less than 100 bp.By gradient diluting the standard viral RNA and detection by one-step SYBR Green RT-qPCR,the best primers were selected from the index of the melting curve and amplification efficiency.The sequences of the primers were showed in Table 1.We found that Q3 primer had ideal amplification efficiency and were more specific than the others(Figure 1).So Q3 RNA was used as standard RNA in the next experiments(Table 2).
2 Specificity and sensitivity of SYBR Green RT-qPCR for M 1 standard RNA detection
RT-qPCR was performed to confirm the specificity and sensitivity of the Q3 primer.According to the RNA concentration,RNA copies were calculated and diluted into 1×1015copies/L.In order to simulate the real situa-tion of RNA extraction from the tissues in vivo,the RNA from different sources,including serum,feces,urine and liver,as matrix RNA was added into the standard RNA.As the RNAs from the liver are rich of RN-ases which can mimic the most stringent condition among all tissues,we first used liver RNA as matrix RNA.Gradient diluted Q3 standard RNA(from 8×107to 80 copies)groups were added into 1 μg matrix RNA(extracted from the liver of SD rats or cynomolgus monkeys),followed by one-step SYBR Green RT-qPCR.The strong matrix effects(samples without standard RNA were also be amplified by the primer,see Figure 2A and 2B)either in SD rats or in cynomolgus monkeys in low concentration group were observed.Using the liver RNA of SDrats as matrix,the standard RNA of 80 copies group was undetectable(Figure 2A).The R2of the standard curve was 0.935,the slope was-3.026,and the Cq values of 80 copies group and control(blank matrix RNA)group were 30.56 and 30.79,respectively.A similar result from using the liver RNA of cynomolgus monkeys was also obtained.Consistently,the standard RNA of 80 copies group and 800 copies group was undetectable in the presence of cynomolgus monkey liver RNA as a matrix(Figure 2B).The R2of the standard curve was 0.923,the slope was-2.906,and the Cq values of 80 copies group and control group were 29.06 and 29.02,respectively.Both results indicated an insufficient sensitivity of the Q3 primer for M1 virusquantification.
To improve the specificity and sensitivity of the Q3 primer,we optimized the experimental conditions and explored the effects of different annealing temperatures on the cynomolgus monkey liver RNA as matrix RNA,and the gradient temperature exploration of annealing temperature showed that above 62℃reduced the matrix effect.Considering that increasing the annealing temperature might affect the amplification efficiency,an annealing temperature of 62℃was chosen to repeat the experiment.As clinical trial recommends that the minimum detection value of standard curve is 50 copies,we added a group of 40 copies into 1μg matrix RNA as a minimum group.The result showed that increased annealing temperature reduced the matrix effect,but the viral RNA in 40 copies group was not detected,indicating that the SYBR Green RT-qPCR was still unable to improve the sensitivity(Figure 2C).Therefore,the SYBR Green RT-qPCR was neither specific nor sensitive enough for M1 virus quantification in tissues.
3 Specificity and sensitivity of TaqMan RT-qPCR for M 1 standard RNA detection
In order to increase the specificity and sensitivity of M1 virus quantification,one-step TaqMan RT-qPCR,which is considered to be more specific than SYBR Green,was tested.Considering the length of Q3 product was 90 bp,which was closed to the standard RNA Q3,and may not be an ideal binding site,resulting in high non-specificity.In order to improve the specificity of the primer,we redesigned a pair of Q3 short primers(Q3S)whose product was 74 bp.In this way,Q3S was better paired with standard RNA Q3.According to Q3 and Q3Sprimers,we designed Q3 and Q3Sprobes for TaqMan RT-qPCR(see Table 3).In order to verify the availability of Q3Sprimer,the amplification efficiency of Q3 and Q3Sprimers using cynomolgus monkey liver RNA as matrix was compared.We found that no fluorescence was detected in 50 copies,100 copies and 800 copies using Q3 primer(Figure 3A).In opposite,using Q3Sprimer,the Cq value of 50 copies group was 36.14.The amplification efficiency of Q3S was 90.2%.The R2was 1.000,and the slope was-3.582(Figure 3B).Therefore,the Q3S primer possessed the ability to detect a low level of viral standard RNA in an interference condition with liver matrix RNA in the TaqMan-based RT-qPCRassay.
We next determined the specificity and sensitivity of TaqMan RT-qPCR using Q3Sprimer to detect the Q3 standard RNA in liver matrix from SD rats and cynomolgus monkeys.The result of SD rats(adding liver matrix RNA as mentioned above)showed that the Cq value for 100 copies group was 37.63,the amplification efficiency of Q3S was 80.0%,the R2was 0.993,and the slope was-3.916(Figure 3C).To verify the accuracy of the standard curve,unknown different dilutions of Q3 standard RNA were also detected as the unknown points,and they were all attached to the standard curve.The accuracy of different values was between 50%~200%,and the coefficient of variation(CV)of 5 repeated results was under 15%.The sample stability test showed that the recovery rate was between 50%and 200%,and the method still maintained a high recovery rate after being stored for 114 d under the condition of-70℃,but the data indicated that repeated freezing and thawing of the samples should be avoided.In addition to extracting matrix RNA from liver,the detection efficiency of this method in the context of matrix RNA from other tissues including excrement,urine and serum was compared,and the accuracy was all between 50%~200%.These results further supported the superior specificity and sensitivity of TaqMan RT-qPCR for M1 virusdetection and quantification.

Figure 2.Matrix effect in SYBR green RT-qPCR using liver RNA from SD rats or cynomolgus monkey as matrix.A and B:gradient diluted standard viral RNA added in 1 μg matrix RNA of SDrat liver(A)or cynomolgus monkey liver(B)detected by one-step SYBRGreen RT-qPCR.The standard RNA of 80 copies group and 800 copies group was undetectable,indicating the insufficient sensitivity.C:using an annealing temperature of 62℃ to detect the standard RNA mixed with cynomolgus monkey liver RNA in one-step SYBRGreen RT-qPCRassay,resulting in the reduced matrix effect.
We have previously demonstrated that the M1 virus was safe in nonhuman primates because no toxicity was observed in the general clinical observations,chemical tests, immunological tests,imaging, and pathological examinations[11].Therefore,it is of great significance to verify whether the detection method can be used for nonhuman primates in the future non-clinical study.The results in cynomolgus monkeys were consistent with those in SD rats.In detecting Q3 standard RNA mixed with liver matrix RNA,the Cq value for 100 copies group was 33.65,the amplification efficiency of Q3Swas 100.0%,the R2was 1.000,and the slope was-3.322(Figure 3D).Similar to the results of SD rats,the unknown points were also attached to the standard curve.The accuracy of different values was between 80%~120%,and CV of 5 repeated results was under 10%.The sample stability test also showed an acceptable recovery rate between 50%and 200%,and the method still maintained a high recovery rate after being stored for 138 d under the condition of-70℃,but the data indicated that repeated freezing and thawing of the samples should be avoided.The experiments mixed with other matrix RNA including excrement,urine and serum also showed good specificity and sensitivity as the accuracy was all between 50%~200%.Collectively,these results confirmed that the TaqMan-based one-step RT-qPCR had sufficient specificity and sensitivity for the detection of M1 virus in the background of matrix RNA.For more details about SYBR Green RT-qPCR and TaqMan RT-qPCR in SD rats and cynomolgus monkeys,please contact us and it is our commitment to provide this information.
4 Tissue distribution of M 1 virus in vivo detected using TaqMan RT-qPCR
To verify whether this assay can be used to detect the tissue distribution of M1 virus,the in vivo experiments in SD rats and cynomolgus monkeys were conducted.Normal SD rats and cynomolgus monkeys were intravenously injected with M1 virus for 3 cycles,and then the M1 viral loads in various tissues,including heart,liver,lung,kidney and brain,were detected.The results showed that the RNA copies of M1 virus in different tissues were quantified(Figure 4A and 4D),which means the novel TaqMan-based one-step RT-qPCR was suitable for quantifying M1 virus in vivo.As the results showed,after multiple rounds of intravenous injections of high-titer(>107CCID50/animal)M1 virus,the M1 viral load on day 44 in each tissue of SD rats was between 100 copies and 800 copies,which was a very low viral load.In cynomolgus monkeys,the viral load on day 44 was lower,which was basically below 200 copies in each tissue,and the value of brain was below the lower quantification limit.The lowest viral load could be detected in the liver,which content was only 28.8 copies.On day 72,no virus was detected from various tissues either in SD rats or in cynomolgus monkeys.These results suggested that the content of M1 virus in normal tissues was extremely low,and the virus did not accumulate in vivo over time but cleared eventually.Moreover,intravenous injection of M1 virus did not affect the weight and temperature of the SD rats and cynomolgus monkeys(Figure 4B,4C,4E and 4F).In addition,after multiple rounds of M1 intravenous injection,no pathological damage was found in the major organs including heart,liver,lung,kidney and brain both in SD rats and cynomolgus monkeys.These results indicated that M1 virus did not accumulate or cause pathological damage in normal tissues,thus explaining the safety of this virus to a certain extent.
We next used TaqMan RT-qPCR to quantitatively detect the M1 viral load in tumor-bearing nude mice at different time points.The M1 virus was intravenously injected into Hep3B tumor-bearing BALB/c nude mice in a single dose of 6.75×109CCID50/kg.Different from other tissues,the viral load gradually increased in the tumor,peaked at 48 h after administration,and then gradually decreased over time.Until 672 h after administration,the M1 viral RNA was still detected in the tumor(Figure 4G).In contrast,the viral load in normal tissues was high within 2~24 h from the beginning of the administration and gradually decreased thereafter,and only a small amount of virus was detected until day 28.Furthermore,the viral load in serum was the highest at 2 h after M1 injection,and then gradually decreased.On day 28,only one serum sample was detected as viral RNA positive.These results further supported that the oncolytic virus M1 targets tumor for efficient oncolysis.
DISCUSSION
In 2014,we reported alphavirus M1 was a selective oncolytic virus targeting ZAP-defective human cancers[10].Moreover,we also found several synergists that greatly enhanced the oncolytic effect of M1 virus[12-14].With the further research on M1 virus,we exerted ourselves to develop M1 virus into a clinically anticancer agent.However,before oncolytic M1 actually carrying out clinical trials,we need to establish the method for viral quantification.

Figure 3.The specificity and sensitivity of TaqMan RT-qPCR for M1 standard RNA detection.A and B:one-step TaqMan RT-qPCRusing Q3 primer(A)or Q3Sprimer(B)for detection of M1 standard RNA mixed with liver matrix RNA of cynomolgus monkeys;C:using one-step TaqMan RT-qPCR to detect M1 viral standard RNA mixed with liver matrix RNA of SD rats;D:using one-step TaqMan RT-qPCR to detect M1 viral standard RNA mixed with liver matrix RNA of cynomolgus monkeys.

Figure 4.Distribution of the number of viral RNA copies in the tissues and the pathological changes from M 1-infected host.A:M1 viral quantification on day 44(n=10)and day 72(n=5)of different tissues in SD rats using one-step TaqMan RT-qPCR;B and C:body weight(B)and temperature(C)of SDrats intravenously injected with or without M1 virus;D:M1 viral quantification on day 44(n=3)and day 72(n=3)of different tissues in cynomolgus monkeys using one-step TaqMan RT-qPCR;E and F:body weight(E)and temperature(F)of cynomolgus monkeys i.v.injected or non-injected with M1 virus;G:total RNAs of different tissues from BALB/c nude mice were extracted at the indicated time points,and the M1 viral load was quantified by one-step TaqMan RT-qPCR(n=6).N.D.:not detectable;n.s.:not significant.Mean±SD.**P<0.01 vs0 h group.
Here,we describe a method of TaqMan probebased one-step RT-qPCR to specifically and sensitively quantify M1 viral RNA.This method detected at least 50 copies of viral RNA in 1μg matrix RNA which is sensitive enough to detect clinical samples.Compared with TaqMan RT-qPCR,the minimum limit using onestep SYBR Green RT-qPCR was only 800 copies in 1 μg matrix RNA.In addition,using SYBR Green RT-qPCR with matrix RNA only,which has no viral RNA,also appeared fluorescence.In a word,the TaqMan probe-based one-step RT-qPCR is sensitive enough to meet the requirement of accurately detecting the quantity of M1 virus and controlling the dosage of M1 virus in clinical application.For clinical trials,a standard curve should be made for quantifying virus RNA.However,as is known to all,it is difficult to preserve RNA for a long time because of the ubiquitous RNA enzymes.Therefore,we used nucleic acid synthesis technique to synthesize a viral RNA fragmentin vitroas the standards.First,according to the sequence of M1 virus,we designed SYBR Green RT-qPCR primers by Beacon Designer and found that the Q3 primer was spe-cific enough to detect M1 virus,so we used Q3 as the best primer for experiment.Due to the instability of viral RNA,it cannot be used as a standard RNA in clinical trials,so we synthesized Q3 standard RNA in vitro according to the product sequence of Q3 primer.However,when we used standard Q3 as template RNA,we found a strong matrix effect in the group of low concentration.Even if optimizing the conditions,such as improving annealing temperature and increasing primer concentration,the matrix effect still existed.It may be because the size of Q3 standard RNA was close to the product size of Q3 primer,and the Q3 primer had no enough position to complementarily pair with the Q3 standard RNA and did not normally recognize the specific binding site of Q3 standard RNA.To solve these problems,we designed the TaqMan RT-qPCR primers and probes of Q3 standard RNA,and found that Taq-Man RT-qPCR effectively improved the specificity of Q3 standard RNA.In order to improve the amplification efficiency and specificity,we also designed a new Q3 short primer(Q3S)to make a better base complementary pairing with Q3 standard RNA.
Clinical trials recommend that the minimum detection value of standard curve is 50 copies.When we used SYBRGreen RT-qPCR,the Cq value of blank matrix RNA was very close to that of the low-concentration standard RNA,so the experiment did not reach the minimum detection limit.Later when we used TaqMan RT-qPCR,no fluorescence was detected in the blank matrix,and 50 copies of the viral RNA in 1μg of standard RNA was detected,indicating that the matrix effect was greatly reduced.So the TaqMan RT-qPCR is sensitive and specific enough to reach the minimum detection value.This may be an ideal method for the lower detection limit of oncolytic RNA virus and for the quantitative detection of other RNA virus.
Viruses are pathogens that strictly depend on their host for propagation.In vivo studies of M1 viral distribution is of great importance to reveal the M1 virus targeting and safety.Our previous researches reported that M1 was not enriched in other normal tissues but was more than 1 000 times enriched in tumor,suggesting the targeting of M1 virus[10].However,we did not absolutely quantify the distribution of M1 virus in other normal tissues.When clinical trials are actually carried out,we must confirm the absolute load of M1 virus to prevent pathological changes caused by excessive load of M1 virus,so as to ensure the safety of M1 virus in antitumor therapy.Our results proved that the Taq-Man RT-qPCR was verified to be satisfactory,as the RNA copies of M1 virus were detected in various tissues of different animals.In addition,these results also indicated that the M1 virus mainly replicated in tumor tissue and did not accumulate in normal tissues.The clearance of virus from body may be due to activation of the body's antiviral immune system,e.g.production of neutralizing antibody.We reported that M1 virus was nonpathogenic for the nonhuman primates after multiple rounds of intravenous injections[11],and we now prove that viral load in various tissues of cynomolgus monkeys was basically below 200 copies in each tissue,which explains that M1 virus will not cause pathological damage and suggests a natural safety of M1 virus as an oncolytic virus candidate.
In conclusion,the TaqMan-based one-step RT-qPCR constructed with Q3Sprimer can be used for M1 virus quantification in various tissue samples of different animals with specificity,efficiency and sensitivity.Therefore,this method will be suitable for further clinical use and help to advance the progress of clinical safety testing of M1 virus.
