Methane production from rice straw carbon in five different methanogenic rice soils: rates, quantities and microbial communities
2020-05-06····
· · · ·
Abstract The input of organic substances(e.g.,rice straw)in rice field soils usually stimulates the production and emission of the greenhouse gas methane (CH4). However,the amount of CH4 derived from the applied rice straw, as well as the response of bacterial and archaeal communities during the methanogenic phase, are poorly understood for different rice field soils. In this study, samples of five different rice soils were amended with 13C-labeled rice straw (RS) under methanogenic conditions. Immediately after RS addition, the RS-derived CH4 production rates were higher in soils (Uruguay, Fuyang) that possessed a stronger inherent CH4 production potential compared with other soils with lower inherent potentials (Changsha, the Philippines, Vercelli). However, soils with higher inherent potential did not necessarily produce higher amounts of CH4 from the RS applied, or vice versa. Quantitative PCR showed copy numbers of both bacteria and methanogens increased in straw-amended soils. High-throughput sequencing of 16S rRNA genes showed distinct bacterial communities among the unamended soil samples, which also changed differently in response to RS addition. Nevertheless,RS addition generally resulted in all the rice field soils in a relative increase of primary fermenters belonging to Anaerolineaceae and Ruminococcaceae.Meanwhile,RS addition also generally resulted in a relative increase of Methanosarcinaceae and/or Methanocellaceae.Our results suggest that after RS addition the total amounts of RSderived CH4 are distinct in different rice field soils under methanogenic conditions. Meanwhile, there are potential core bacterial populations that are often involved in primary fermentation of RS under methanogenic conditions.
Keywords 13C-labeled rice straw · Methane production ·Rice field soil · Microbial community
1 Introduction
The plant organic substances, such as rice straw, are often applied in large amounts(up to 12 Mg/ha annually)to rice field soil so as to maintain soil fertility (Liu et al. 2014;Sass et al. 1991; Schu¨tz et al. 1989; Yagi and Minami 1990).Rice straw is typically added at the beginning of the season before rice planting,but other types of plant organic matter are continuously added during the rice-growing season since organic substances such as root exudates,sloughed-off leaves,and roots are released into soils during the life of rice plants (Dannenberg and Conrad 1999; Lu et al.2002).Such organic matter input eventually results in a steady increase of soil organic matter in rice field soils(Kalbitz et al. 2013; Liu et al. 2006, 2014 Mitra et al.2005).However,incorporation of plant organic matter also increases the production and emission of CH4, a potent greenhouse gas (Denier van der Gon and Neue 1995; Gao et al. 2016; Haque et al. 2013; Kim et al. 2012, 2015;Kimura et al. 2004; Schu¨tz et al. 1989; Yagi and Minami 1990).The contribution of rice fields to atmospheric CH4is about 10%(Conrad 2009).Methane is produced during the anaerobic decomposition of organic substances by bacterial populations involved in hydrolysis, fermentation, syntrophy and homoacetogenesis together with methanogenic archaea (Conrad 1999; Schink and Stams 2013). The methane production capacity of the rice field soil without organic amendment was considered as its inherent methane production potential (Lu et al. 2000), which was mainly controlled by the availability of degradable organic substrates in soil (Yao and Conrad 1999; Yao et al. 1999).
The more rice straw is added to rice field soil,the higher is the production of CH4(Bao et al. 2014; Denier van der Gon and Neue 1995; Naser et al. 2007). Meanwhile, the addition of organic substances,including root exudates and rice straw seems to trigger a faster and higher response of CH4production rates when the soils possess a high inherent CH4production potential compared to those with low production potential (Lu et al. 2000). This is consistent with the observation that identical amounts of straw incorporation can cause different timing and different shapes of emission peaks of CH4in different rice field soils(Yagi and Minami 1990). There are numerous studies of the influence of RS addition on the methanogenic archaeal communities, concluding that members of these communities respond differently to the type of organic residues and incubation temperature and duration (Conrad and Klose 2006;Conrad et al.2012;Lu et al.2015;Peng et al.2008). For example, the incorporation of rice straw selectively enhanced the growth of Methanosarcinaceae and suppressed rice cluster I (Methanocellaceae) methanogens(Conrad and Klose 2006). However, the response of the bacterial community to rice straw (RS) addition has only rarely been investigated (Ji et al. 2018; Rui et al. 2009;Wegner and Liesack 2016).
Little attention has been paid to the total amount of CH4that is derived from the plant organic substances during the methanogenic phase in different rice soils, although it is very important for the evaluation of the greenhouse gas emissions by adding organic matter in rice field soils.Rice straw,which mainly consists of polysaccharides plus some lignin, is relatively easily degradable, so that after one season typically 80–90% of the added rice straw has disappeared (Neue and Scharpenseel 1987). Therefore, a significant percentage of rice straw should be converted to CH4and CO2during several weeks of incubation. However,CH4is produced not only from the degradation of rice straw but also from soil organic matter and root exudates(Kimura et al. 2004). For quantification of the RS-derived CH4, it is, therefore, better to measure the CH4produced from isotopically labeled RS (Yuan et al. 2012a). In addition, at the beginning of flooding (fresh, non-methanogenic soils) there will be a succession of bacterial and archaeal communities along with the sequential reduction of inorganic electron acceptors (including SO42-and Fe(III)) (Lueders and Friedrich 2000; Yao et al. 1999;Yuan et al. 2012a; Yuan and Lu 2009). Later on, when methanogenic conditions are established the microbial community in rice field soil becomes relatively stable (Yuan et al. 2012a; Yuan and Lu 2009). In order to account for the methanogenic conversion of RS, straw should preferentially be added when the methanogenic community has already established. The responses of the microbial communities to RS addition might then be different from that at the beginning of flooding. In-depth knowledge of the amount of plant organic substancesderived CH4production and the response of the microbial community to plant organic substances is of particular importance for the development of options to mitigate CH4emissions.
In the present study, five flooded rice soil samples were amended with13C-labelled rice straw after preincubation for 40 days under anaerobic conditions, so as to (1)determine the amount of RS-derived CH4and CO2production, and (2) identify the response of bacterial and archaeal community in different soils after RS amendment under methanogenic conditions. We hypothesized that in different rice field soils, the total amounts of RS-derived CH4are different and there are core microbial populations involved in the decomposition of RS under methanogenic conditions.
2 Materials and methods
2.1 Soil samples
Five different paddy soils were collected from China(Fuyang and Changsha), the Philippines, Italy (Vercelli)and Uruguay. The Fuyang soil, a clay loam, was collected from a rice field at the China National Rice Research Institute in Hangzhou(Rui et al.2009).The Changsha soil,a sandy clay loam, was collected in a typical double rice cropping field of Changsha County in Hunan Province,China. Uruguay soil, a silt clay soil, was sampled from a field 70 km from the Instituto Nacional de Investigacio´n Agropecuaria (INIA) at the city Treinta-y-Tres, Uruguay(Fernandez Scavino et al. 2013). The Vercelli soil is a silt loam that originated from a rice field at the Italian Rice Research Institute in Vercelli, Italy (Yuan et al. 2012b).The Philippine’s rice field soil is a silt loam collected at the International Rice Research Institute in Los Banos, the Philippines (Breidenbach and Conrad 2015). The rice soil samples were air-dried, sieved (<0.5 mm) and stored at room temperature (Ma et al. 2010). The main characteristics are given in Table 1.The storage of dry soil samples at room temperature does not significantly influence its CH4production capacity (Mayer and Conrad 1990).
2.2 Soil incubation
For the soil incubation,4 g dry weight soil was mixed with 6 ml anoxic water in each 26-ml pressure tube as described before (Yuan et al. 2014). Each tube was sealed with a butyl rubber stopper together with aluminum crimp, then flushed with pure N2for 3 min and incubated statically at 25 °C under dark condition. The production of CH4, CO2and Fe(II)were measured during the incubation(Yao et al.1999). After 40 days of preincubation, during which any available ferric iron or sulfate was reduced, each tube was amended with 8 mg(0.2%)of RSI or RSII powder so as to prepare RS treatments I and II.An unamended control was run in parallel. The amount of 8 mg RS is equivalent to 267 μmol C. The application of 0.2% RS equals field conditions when the rice straw is applied at roughly 3.6 Mg/ha (considering soil bulk density is around 1.2 g/cm3). The δ13C values of RSI (596.1‰) and RSII(885.0‰) were achieved by mixing the desired amount of13C-labeled (δ13C = 1859.9‰) and unlabeled (δ13-C = - 27.6‰) RS. All the RS originated from rice plants cultivated in the greenhouse,13C-labeled RS was obtained by cultivating the rice plants in the chamber amended with13CO2(Yuan et al. 2012b). The rice plants were harvested at the late vegetative stage,then dried at 60 °C and ground to powder. The tubes were sealed again and flushed with N2immediately after amended with RS,then shook and reflushed with N2so as to remove the residual CH4and CO2.At last, the tubes were incubated under the conditions as described above for 22 days. Triplicates were prepared for all the treatments.
2.3 CH4 and CO2 analyses
The analyses of CH4and CO2were the same as described before (Yuan et al. 2018). In brief, every 2 to 5 days, gas samples taken with gas-tight syringe from the headspace of the tubes were injected into a gas chromatograph (GC)equipped with flame ionization detector (FID), so as to analyze the concentrations of CH4and CO2(Bodelier et al.2000). The CO2was measured after conversion to CH4with a methanizer(nickel catalyst at 350 °C)(Penning and Conrad 2007). At the end of incubation, the liquid was analyzed for pH after the tubes were opened. Total inorganic carbon (TIC) was defined as the sum of gaseous,dissolved and bicarbonate CO2(Stumm and Morgan 1981)and determined at the end of incubation.
The δ13C values of CH4and CO2were determined using gas chromatography combustion isotope ratio mass spectrometry (GC-C-IRMS) (Finnigan, Bremen, Germany), as described in an earlier study (Penning and Conrad 2007).The δ13C values of gaseous and bicarbonate CO2were calculated from the δ13C of gaseous CO2based on the corresponding fractionation factors (Stumm and Morgan 1981),all these δ13C values were used to calculate δ13CTICaccording to the mole fractions of each CO2species(Penning and Conrad 2006).
2.4 Calculation of CH4 and CO2 derived from the RS applied
The calculations of CH4and CO2derived from the RS applied were the same as described earlier (Yuan et al.2014;2018).In brief,since the treatments RSI and RSII are only different in the δ13C of the RS, the fraction of CH4derived from RS (fRS) could be calculated by:

of which the δ13C values were measured during the incubation experiment. The δ13CCH4-Iand δ13CCH4-IIwere the δ13C values of the CH4produced in the tubes of RS treatment I and II,respectively;the δ13CRS-I(596.1‰)andδ13CRS-II(885.0‰)are δ13C values of the RS carbon in RS treatment I and II, respectively.

Table 1 Characteristics of the soil samples
Next,the amount of CH4derived from RS(pRS,CH4)was calculated from the fRSand the total amount of CH4produced (pCH4) in the tube:

Analogous equations are valid for calculation of the fractions and amounts of CO2derived from RS.
2.5 DNA extraction of soil samples and quantification of microbial abundance
The soil samples collected after the incubation were used for DNA extraction with the FastDNA® Spin kit for soil(Qbiogene, Germany). For the quantifications of copy numbers of bacterial 16S rRNA, the primer pair (519f and 907r) and parameters were applied according to the protocol described before (Stubner 2002); for mcrA gene,which is marker gene for methanogenic archaea,the primer pair (mlas-mod and mcrA-rev) and parameters were performed according to the protocol described in(Angel et al.2011). The quantitative PCR was done on iCycler thermocycler (Bio-Rad, Germany).
2.6 Pyrosequencing of the bacterial and archaeal communities
Tagged pyrosequencing of the bacterial and archaeal 16S rRNA genes was performed using primer pairs F515/R806(Bates et al. 2011) and Arch344F/Arch915, respectively(Casamayor et al. 2002; Yu et al. 2008). The forward primer of each pair contained a unique 6-bp barcode (Hernandez et al. 2015). Sequencing of the pooled PCR products was carried out at the Max Planck Genome Centre in Cologne with a Roche 454 Genome Sequencer GS FLX+.
The raw sequence data were analyzed with Mothur (v.1.27) software package (http://www.mothur.org/) (Schloss et al.2009).Sequences were removed from further analysis in case they were less than 200 bp, harbored ambiguous bases or homopolymers longer than 6 bp. Chimeras were removed with the UCHIME (Edgar et al. 2011). 97%sequence similarity was used to define the operational taxonomic unit (OTU) from the good sequences. OTU table was created using the UPARSE pipeline (Edgar 2013). Rarefaction, Shannon index, and Chao1 estimators of OTUs were calculated in RDP(http://pyro.cme.msu.edu/). The phylogenetic affiliation of each bacterial and archaeal 16S rRNA gene sequence was analyzed with the RDP Classifier at a confidence level of 80%.Resampling to the same sequence depth was conducted before the downstream analysis. The original pyrosequencing data were deposited in the NCBI Sequence Read Archive(SRA)under the study number SRP058834.
2.7 Statistical analysis
The redundancy analysis(RDA)was performed to measure the influence of environmental variables on bacterial and archaeal communities using the R package Vegan (http://cran.r-project.org/web/packages/vegan/index.html). Linear discriminant analysis (LDA) effect size (LEfSe) was used to detect bacterial taxa differentiating the different rice soil samples(Segata et al.2011).Two-tailed independent t test and one-way analysis of variance (ANOVA) were used to test the significance of the differences between and among the treatments on various variables, respectively.
3 Results
3.1 CH4 production rates in control and RSamended rice field soils
Anoxic incubation of soils with RS and without (control)resulted in linear accumulation of CH4with time.Averaged CH4production rates were calculated from the total amount of CH4accumulated during the whole incubation period(22 days)in control and RS treatment.The results of the control soils showed that Uruguay soil had the highest inherent potential for CH4production,and Fuyang soil also had apparently higher CH4production potential compared with other soils (Fig. 1a). The RS addition substantially stimulated the CH4production rates in all soil samples.After RS addition, the CH4production rates in these soil samples were 2 to 6 times of those in the control.
3.2 RS-derived CH4 and CO2 production in different rice field soils
The accumulated total amounts of CH4derived from the added RS were calculated with Eq. 2. On day 5 after RS addition, the amounts of RS-derived CH4in Uruguay and Fuyang soil were about 3 and 2 times, respectively, larger than those in the other three soils (Fig. 1b). In most soils,the accumulated amounts of RS-derived CH4reached the maximum at day 15.In Vercelli soil, however,the amount of RS-derived CH4still increased steadily afterward. The total amounts of RS-derived CH4were in a range of 35–55 μmol (Fig. 1b). Eventually, the amounts of RSderived CH4in Vercelli and Uruguay soils were by 21 to 46% higher (P <0.01) than those in the other soils. The total amounts of RS-derived CO2(shown as TIC) were around 80 μmol at the end of incubation (Fig. 1c). They were relatively lower in Changsha soil than in the other soils. The total amounts of RS carbon mineralized to CH4+CO2in soil from Changsha, Fuyang, the Philippines, Uruguay and Vercelli were 115.7 ± 2.6,116.5 ± 10.2, 125.3 ± 5.7, 133.5 ± 11.8, and 134.5 ± 3.1 μmol, respectively. These amounts are equivalent to 42–54% of the added RS carbon.

Fig. 1 Averaged CH4 production rate in control and rice straw(RS)treatment of each soil sample(a).Accumulation of RS-derived CH4 in each soil sample amended with RS(b).Amounts of RS-derived total inorganic carbon(TIC)in RS amended soil samples at the end of incubation(day 22)(c).The data shown are from treatment RSI since they were almost identical to the results from treatment RSII.Data are mean ± SD(n = 3).The differences between different soil samples were tested by one-way analysis of variance (ANOVA), different letter on the top of the bar between the column indicated a significant difference (P <0.05) between the soils
3.3 Abundance and composition of bacterial and archaeal communities in control and RSamended rice field soils
The results of quantitative PCR showed that in unamended control soils, the copy numbers of bacteria and methanogenic archaea ranged from 4.4 × 109to 9.0 × 109and from 2.5 × 107to 1.3 × 108,respectively.RS addition led to a significant increase in the abundance of bacteria only in Fuyang and Philippines soils, while the abundance of methanogenic archaea significantly increased in almost all soils after the RS amendment (Table 2). The final ratio between the mcrA gene and bacterial 16S rRNA gene copynumbers (Fig. 2) was lower in most of the control soils(0.005–0.01) than in Fuyang soil (0.023). After RS addition, however, this ratio decreased in Fuyang soil while it substantially increased in the other soils.
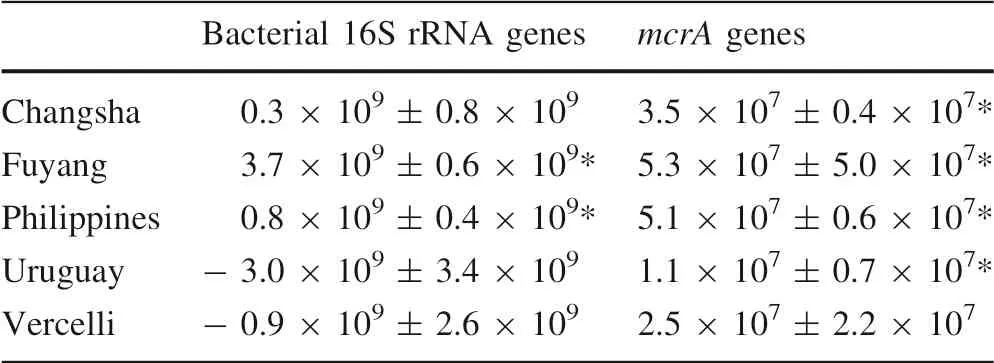
Table 2 Increase of copy numbers of bacterial 16S rRNA gene and mcrA gene in each soil given by the difference before and after RS addition; mean ± SD (n = 3)
Pyrosequencing analysis of bacterial 16S rRNA genes indicated that Acidobacteria, Actinobacteria, Clostridia,Chloroflexi, and Deltaproteobacteria were the dominant bacterial groups in each soil sample (Fig. 3a). Among them, the relative abundance of Clostridia was obviously higher in Fuyang soil than in the other soil samples. After RS addition, the relative abundance of Clostridia apparently increased in the soils from Uruguay, the Philippines and Changsha. The relative abundance of Chloroflexi was elevated in Changsha, Fuyang and Vercelli soils after RS addition.
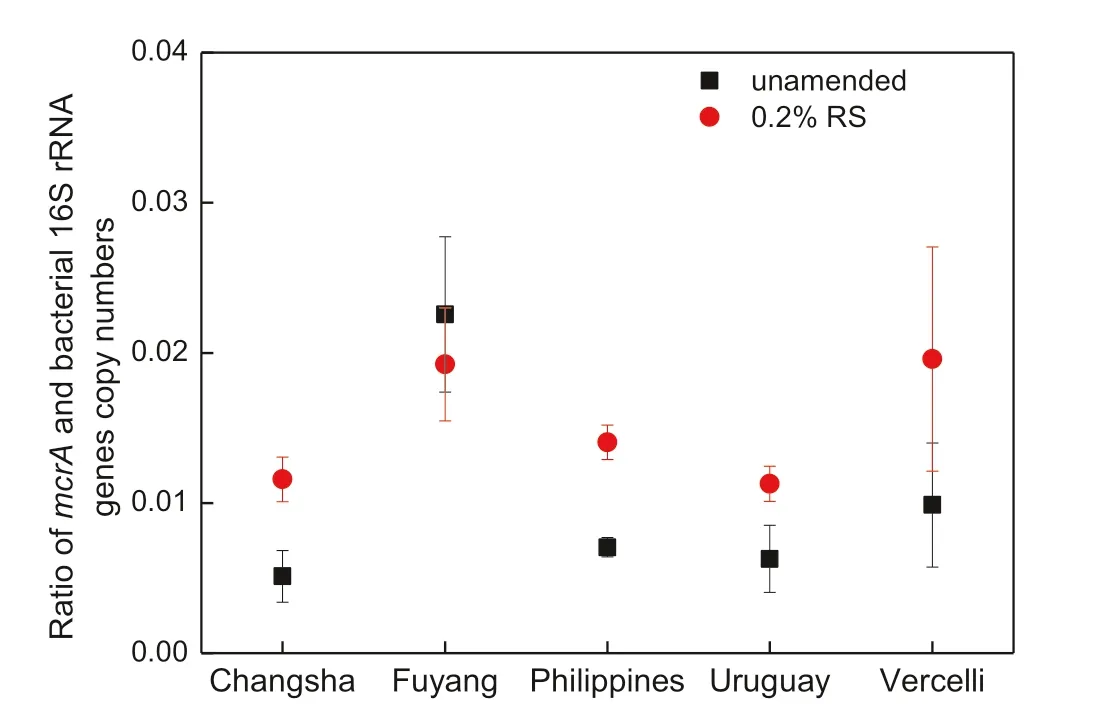
Fig. 2 Ratio between mcrA gene and bacterial 16S rRNA gene copy numbers in soils amended and unamended with RS at the end of incubation (day 22); mean ± SD (n = 3). Results of Fuyang and Uruguay soils were calculated from data presented in Yuan et al.2018

Fig. 3 Relative abundances of bacterial phyla or classes (a), and archaeal families (b) in control and RS treatment of each soil sample. Major taxa with average relative sequence abundances higher than 1%are displayed;mean ± SD(n = 3).Relative abundances of all the rare phyla or families(<1%in all samples),candidate divisions and unclassified sequences were combined and indicated with the column‘‘Bacteria-others’’or ‘Archaea-others’’
Sequencing of archaeal 16S rRNA genes indicated that the Methanosarcinaceae and Methanocellaceae were the dominant archaeal families in each soil sample (Fig. 3b).The relative abundance of Methanocellaceae was slightly elevated in Fuyang, Changsha and the Philippines soil samples, while Methanosarcinaceae were substantially higher (by 22.6%) in the Uruguay soil sample, and also slightly higher in the other soils compared to the Fuyang soil. Besides, the relative abundances of Nitrososphaeraceae were higher in the control soils from the Philippines and Uruguay compared with the other soils but apparently decreased in both soils after RS addition.
Redundancy analysis (RDA) was conducted to analyze the relationship between environmental variables and microbial communities. The RDA ordination showed that specific environmental conditions shaped the variations in bacterial and archaeal community compositions (Fig. 4).The bacterial community of Fuyang soil was strongly affected by both soil organic carbon (SOC) and total nitrogen (TN) contents (Fig. 4a). As for the archaeal community, the relatively large vector size indicated that TN was more strongly correlated with the archaeal community composition than with the other tested factors(Fig. 4b).
To detect the bacterial groups that most likely explain the differences among the different soil samples, we used the linear discriminant analysis effect size (LEfSe), a tool for biomarker discovery. LEfSe detected many bacterial groups with statistically significant differences(P <0.001)among the soil samples (Fig. 5). For example, four bacterial families belonging to the class Clostridia were overrepresented in Fuyang soil, while the order Bacillales and the families Beijerinckiaceae and Bradyrhizobiaceae were significantly more abundant only in Uruguay soil. The family Chitinophagaceae and several families affiliated with Deltaproteobacteria, including Geobacteraceae and
Desulfobacteraceae were more abundant in the Philippines soil, but the family Planctomycetaceae was relatively higher in Changsha soil.

Fig. 4 Ordination diagrams from redundancy analysis (RDA) of bacterial (a) and archaeal (b) communities and environmental parameters. Arrows indicate the direction and magnitude of environmental parameters associated with bacterial and archaeal community structures. Each sample is represented by a colored circle (control)and triangle (RS treatment). TN total nitrogen, SOC soil organic carbon

Fig. 5 Taxonomic cladogram obtained from LEfSe analysis of bacterial taxa (relative abundance >0.5% in at least one sample). The LEfSe analysis identified the most differentially(P <0.001)abundant taxa among the soil samples(highlighted by small circles and by shading);color denotes the different soils and that the colored bacterial taxon was significantly overrepresented in the respective soil.The diameter of each dot is proportional to the relative abundance of the taxon.Yellow circles represent non-significant differences in abundance among the soil samples for that particular taxonomic group
3.4 Key bacterial groups responsive to RS addition in rice field soils
In order to identify the key bacterial groups responsive to the addition of RS,we summarized the significant increase of members of the bacterial community at a lower taxonomic rank (Table 3). There were about 17 bacterial families that significantly increased (albeit to different extents) after RS addition, most of them being affiliated with Firmicutes, Acidobacteria, Bacteriodetes, Chloroflexi and Verrucomicrobia.The Uruguay soil exhibited a higher increase (10.8%) in the relative abundance of bacterial families after RS addition than the other soils. This observation was consistent with the increase of several Firmicutes only in Uruguay soil after RS treatment,such as the Clostridiaceae 1 and Clostridiales_Incertae Sedis XVIII. Upon RS addition, there was a general increase in the relative abundances of both Ruminococcaceae and Anaerolineaceae in all soil samples, while Opitutaceae increased to a less extent in most of the soil samples.
4 Discussion
4.1 Methanogenic decomposition of RS in rice field soils with different inherent CH4 production potential
Rice straw typically contains >85% easily degradable organic matter (i.e., soluble material, polysaccharides,lipids) and only low amounts of recalcitrant lignin (about 5–15%) (Watanabe et al. 1993). The decomposition of RS started immediately after RS addition. The slow-down of CH4accumulation after day 15 indicated that the fast phase of RS decomposition was finished probably because the easily degradable organic carbon in RS was exhausted(Lu et al. 2003). A previous study also showed that the decomposition rate of RS was very high in the early stage after RS addition, while the decomposition rate strongly decreased after 22 days (Watanabe et al. 1998). These observations are consistent with the finding that turnover times of cellulose degradation are in a range of 7–14 days in fresh and methanogenic soil (Chidthaisong and Conrad 2000). Since the RS applied was ground to powder for accelerating the decomposition rate, the easily degradable organic carbon of the RS applied was probably almost completely decomposed at the end of the incubation. In Uruguay and Fuyang soil, the apparently higher RS-derived CH4production rates at the beginning of RS decomposition could be attributed to the higher inherent potential for CH4production in both soils compared with other soil samples. Previous studies have reported higher production rates of RS-derived CH4in soils of stronger inherent CH4production potential (Lu et al. 2000).

Table 3 Increase in relative abundances of bacterial families and unclassified bacterial groups in each soil after RS application
However, our results indicated that soils with higher inherent CH4production rates did not necessarily produce higher total amounts of RS-derived CH4or vice versa.The higher total amounts of RS-derived CH4in Uruguay and Vercelli soil samples were consistent with their relative higher total amounts of RS-derived CO2compared with other soil samples (Fig. 1b, c). Thus, methanogenic decomposition of the same amount of RS resulted in more end products(CH4and CO2)in Uruguay and Vercelli soils compared to the other soils. Still, the RS-derived CH4and CO2represented at most 54% of the applied RS carbon.Hence, a large part of the non-recalcitrant RS carbon must have become converted to organic matter that was no longer easily available to methanogenic degradation, perhaps microbial biomass or new soil organic carbon, especially in Fuyang, Changsha and the Philippines soils. This conclusion is consistent with the increase in abundance of bacterial 16S rRNA genes and/or methanogenic mcrA genes that was higher in Fuyang, Changsha and the Philippines soils than in the other soil samples (Table 2).Furthermore, the results suggest that in these three paddy soils straw application may result in a relatively higher elevation of SOC content than in the other soils. This increase possibly also causes higher soil fertility, since increasing SOC content has been found to exhibit a positive correlation with the responses of soil macro-aggregates and crop yield to straw return (Liu et al. 2014).
Meanwhile, the ratios of RS-derived CO2to CH4were between 1.4 and 2.1 in these soil samples, although complete mineralization of polysaccharides (e.g. rice straw)under methanogenic conditions should give equal amounts of CO2and CH4(Conrad et al. 2010; Zinder 1993). This was partly because hydrogenotrophic methanogenesis from SOC-derived CO2was enhanced by H2released during RS decomposition (Yuan et al. 2014) so that the RS-derived CH4production was reduced accordingly. In addition,although acetate carbon was predominantly consumed by acetoclastic methanogens, part of it could also have been assimilated as a C source by a small and heterogeneous community of bacteria,thus resulting in a decrease of CH4production (Hori et al. 2007; Schwarz et al. 2007).
4.2 Responses of bacterial and archaeal communities to RS addition in methanogenic rice field soils
The bacterial communities were apparently different on the phylum level among the different rice field soils (Fig. 3a),being consistent with LEfSE analysis of the different soils(Fig. 5). The differences in bacterial communities were well explained by distinct controlling factors in the different soils, including pH and the contents of iron, soil organic carbon and total nitrogen(Fig. 4).For example,the higher SOC content in Fuyang soil probably explained its obviously higher abundances of bacterial families affiliated with Clostridia, which are usually involved in hydrolysis and fermentation of organic matter(Kim and Liesack 2015;Rui et al. 2015). In addition, the responses of the bacterial communities to RS addition were also largely different among the different soils.
First, there were differential increases of bacterial families affiliated with Acidobacteria, Bacteriodetes, and
Verrucomicrobia among the different soils, although these different taxonomic bacterial groups were probably all involved in hemicellulose breakdown after RS addition(Wegner and Liesack 2016). Moreover, several families affiliated with Clostridia displayed a strong increase after RS addition (Table 3). Among them, Clostridiaceae,Lachnospiraceae etc.were found to predominantly express glycosyl hydrolase transcripts that are involved in cellulose and chitin breakdown after RS addition to Vercelli soil(Wegner and Liesack 2016). However, the relative abundance of Clostridiaceae increased only in Uruguay soil(Table 3), while it decreased in the other soils including Fuyang soil (data are not shown), in which Clostridiaceae had the highest relative abundance (Fig. 5). In contrast, a previous report indicated that RS addition in Fuyang soil at the beginning of flooding largely stimulated the abundance of species affiliated with Clostridiaceae (Rui et al. 2009).These results suggested that the role of Clostridiaceae may change in different soils and under different soil physicochemical conditions.
Common to all the different soils was the observation that Ruminococcaceae and Anaerolineaceae substantially increased. The Ruminococcaceae are known for plant polymer breakdown in gut environments (Jalanka-Tuovinen et al. 2011; Turnbaugh et al. 2009) and rice field soil(Wegner and Liesack 2016).Similarly,most species of the Anaerolineaceae are capable of fermentative metabolism,they could utilize carbohydrates and proteinaceous carbon substrates under anaerobic conditions (Sekiguchi et al.2003;Sun et al.2016;Yamada et al.2006).They are likely important primary fermenters in anaerobic environments,such as anaerobic digester(Mcllroy et al.2017;Meng et al.2017),but their role in the paddy soils was hardly reported previously. Besides, Opitutaceae increased to a less extent in most of the soil samples. The genomic analysis of Opitutaceae bacterium strain isolated from wood-feeding termites showed that it harbors genes coding for lignocellulosic degradation (Isanapong et al. 2012). And OTUs belonging to Opitutaceae were identified consumers of plant-derived carbon in the rice rhizosphere (Breidenbach et al. 2016; Hernandez et al. 2015). On the other hand, RS addition just resulted in a relative increase of
Methanosarcinaceae and/or Methanocellaceae (Fig. 3b),probably because both families were the only dominant methanogenic archaeal groups in each soil samples.
5 Conclusion
Our results suggest that after RS addition the rates of CH4production and the total amounts of RS-derived CH4are distinct in different rice field soils under methanogenic conditions. Two potential core populations consisting of
Ruminococcaceae and Anaerolineaceae were identified,suggesting that they may be indispensable during the primary fermentation of RS under methanogenic conditions.These results are probably important for evaluating the greenhouse effect of rice residues, which will help to improve process-based modeling of CH4emission from flooded rice fields. Meanwhile, these results suggest that the amount of straw return should be reduced in the rice field soils because of its adverse effects on CH4production.
AcknowledgementsWe thank P. Claus and M. Klose for technical assistance, Y.L. Yuan, B. Martin, A. Fernandez Scavino and C.M.Yin for providing soil samples,and J.Pump for providing labeled rice straw. Q.Y. received a fellowship from the Max Planck Society,Germany. This work was supported by the National Natural Science Foundation of China (41573083); by the Construction Program of Biology First-class Discipline in Guizhou (GNYL[2017]009FX1KT09), China; by the LOEWE center for synthetic microbiology(SYNMIKRO),Germany;by the German Research Foundation as part of the ICON consortium (CO 141/4-1).
Compliance with ethical standards
Conflict of interestThe authors declare that they have no conflict of interest.
杂志排行
Acta Geochimica的其它文章
- High oxygen fugacity magma: implication for the destruction of the North China Craton
- The effect of pH on the sorption of gold nanoparticles on illite
- Optimizing the ratio of the spike to sample for isotope dilution analysis: a case study with selenium isotopes
- Antimony removal from wastewater by sulfate-reducing bacteria in a bench-scale upflow anaerobic packed-bed reactor
- Ecological risk assessment of surficial sediment by heavy metals from a submerged archaeology harbor,South Mediterranean Sea,Egypt
- Discrimination geochemical interaction effects on mineralization at the polymetallic Glojeh deposit, NW Iran by interative backward quadratic modeling
