Self-assembly of L-tryptophan on Cu(111)studied by low-temperature scanning tunneling microscopy
2020-01-14QingXueYjieZhngRuoningLiChoLiLiChenyngYunShiminHouYongfengWng
Qing Xue,Yjie Zhng,Ruoning Li,Cho Li,N Li,Chenyng Yun,Shimin Hou,b,*,Yongfeng Wng,c,*
a Key Laboratory for the Physics and Chemistry of Nanodevices, Department of Electronics, Peking University, Beijing 100871, China
b Peking University Information Technology Institute (Tianjin Binhai), Tianjin 300450, China
c Beijing Academy of Quantum Information Sciences, Beijing 100193, China
Keywords:
Scanning tunneling microscopy
Tryptophan
Self-assembly
Surface science
Hexamer
ABSTRACT
The self-assembly of L-tryptophan on Cu(111)is investigated by an ultrahigh vacuum scanning tunneling microscope (STM) at 4.4 K.When deposited onto the substrate at around 120 K with a coverage of 0.1 monolayer, molecular trimers, tetramers, hexamers, and chains coexist on Cu(111).Then almost all molecules self-assemble into chiral hexamers after being annealed at room temperature.When increasing molecular coverage to the full layer,a new type of chain is observed on the surface.Based on the high-resolution STM images at sub-molecular level,we suggest that the L-tryptophan molecules are present in neutral, zwitterionic or anionic states in these structures.
Structures of biomolecules play important roles in functions of living organism.They are usually characterized by X-ray diffraction, nuclear magnetic resonance and cryo-transmission electron microscopy.These methods either average signals from a large number of molecules or can only characterize large biomolecules,like protein.Scanning probe microscopy is a powerful tool to investigate molecules at single-atom level and is capable to invested small biomolecules.Various amino acids such as glycine[1-4], alanine [5-7], serine [8], methionine [9-11]and lysine[12,13]have been intensively studied on surfaces [14].
Tryptophan contains an α-amino group, an α-carboxylic acid group, and an indole functional group (-C8H6N).Therefore,tryptophan molecules can interact with each other through many types of interactions.Its crystal structure is obtained only until 2012 [15].Recently, the adsorption and self-assembly of Ltryptophan molecules on Cu(100), Cu(111), and HOPG have been investigated by scanning tunneling microscopy (STM) [16,17].In these studies, L-tryptophan molecules were evaporated on surfaces which were kept at room temperature.In this case, molecules could diffuse freely on surfaces and formed various ordered patterns.If the substrate is at lower temperature during deposition, the molecular diffusion will be reduced and new molecular structures may be formed.
In this work, we deposit tryptophan molecules on Cu(111) at 120 K and observe three types of molecular clusters and two kinds of chains.Their structures are characterized by STM operated at 5 K with sub-molecular resolution.
The experiments are performed in an ultra-high vacuum STM(base pressure: 5×10-9Pa) at liquid Helium temperature.The single-crystalline Cu(111)substrate is prepared by repeated Ar-ion sputtering at 500 V and annealing at 673 K.The cut Pt/Ir tips are first annealed in vacuum and then softly dipped into Cu(111).LTryptophan(Sigma Aldrich)is thermally sublimated from a homemade Ta boat onto the Cu(111) substrate.During deposition, the sample is held at 120 K.The samples are measured at 5 K and STM images are slightly smoothed using software WSxM.
The amino acids are present in neutral,zwitterionic or anionic states on surfaces[18,19].Compared to the neutral form in the gas phase,amino acids tend to adopt zwitterionic states,with protons transferred from the carboxylic group to the amino group,in solids or in solutions [19,20].Anionic forms are obtained through the deprotonation of carboxylic groups on reactive substrates.In crystal,the L-tryptophan molecule is at the zwitterionic state with a positively charged amino group (R-NH3+) and a negatively charged carboxylate group (R-COO-).It has two types of configurations according to the previous crystallography study[15], which are displayed in Fig.1 and labeled as 1 and 2,respectively.Surface adsorption can generate additional chirality[21,22].Enantiomers δ can be obtained by flipping enantiomers λ with a 180 degree along the direction perpendicular to the substrate.Therefore, there are four types of enantiomers on Cu(111), as shown in Fig.1.Considering the three chemical forms, Ltryptophan molecules adsorb on surfaces in twelve configurations in total, which will be used to interpret the molecular selfassembled structures on Cu(111).
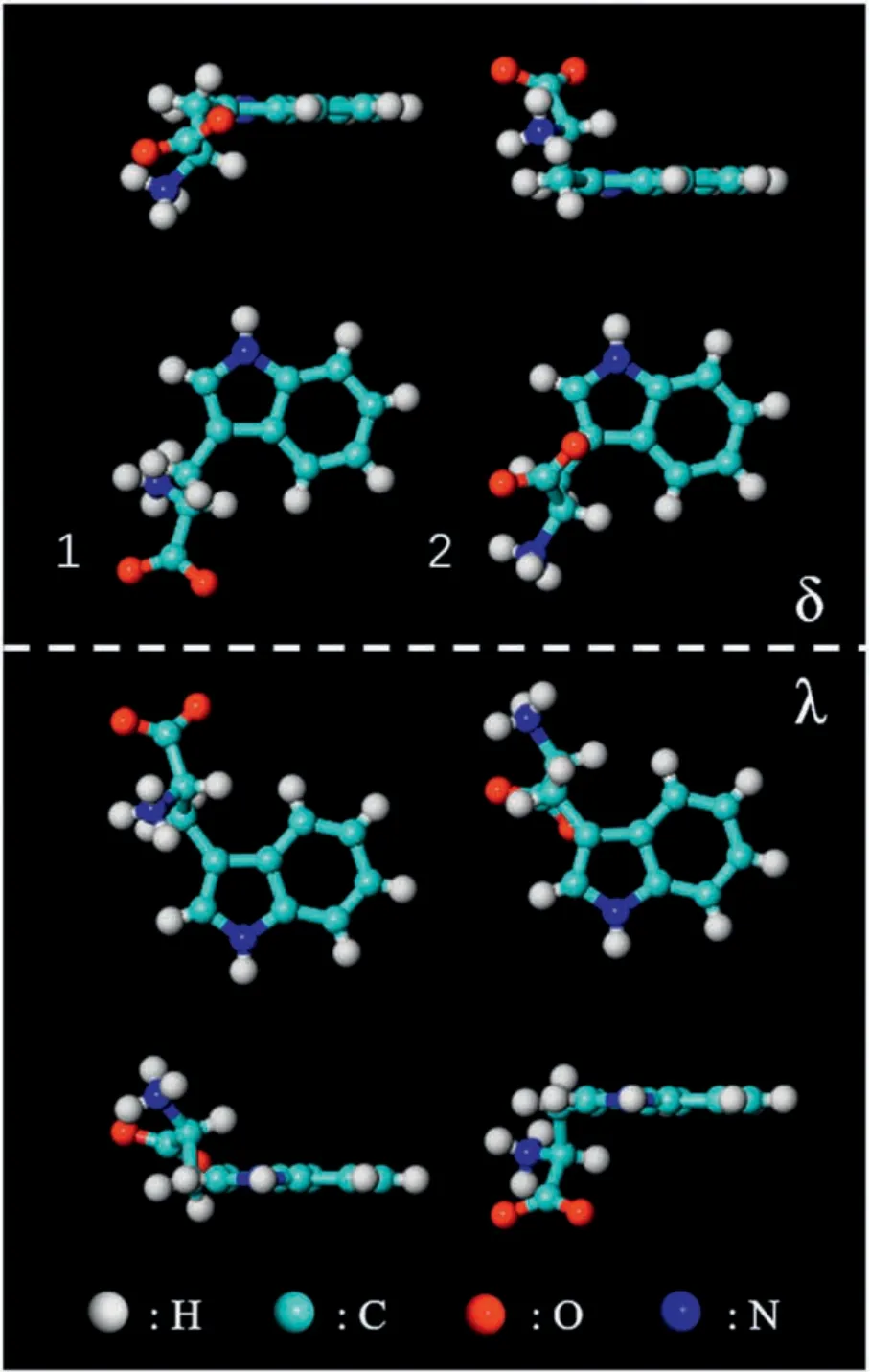
Fig.1.Vertical and lateral views of two configurations of zwitterionic L-tryptophan molecules labeled as 1 and 2.When adsorbed on surface, each of them has two chiral footprints labeled as δ and λ.Color code:cyan(C),white(H),red(O),blue(N).
After the thermal deposition of L-tryptophan and subsequent annealing at 120 K, the Cu(111) surface is covered by a series of different shaped clusters and one-dimensional (1D) dimer chains at a coverage of 0.1 ML, as shown in Fig.2a.The obvious standing waves around the self-assembled structures reveal strong scattering of the surface-state electrons from the molecules [23,24],which prohibits the aggregation of molecular clusters.The dimer chains and clusters emerged alone <1 1> orientations corresponding to 3-fold symmetry of the Cu(111) surface.
To gain further insight, high-resolution STM images of the supramolecular L-tryptophan structures are measured.Detailed structures of a trimer and tetramer are displayed in Figs.2b and d.For molecules with configurations of 1λ and 2δ,the amino and αcarboxylic acid group groups point to vacuum and the indole group lies flat on Cu(111).These two parts should appear differently in STM images.In contrast, 2λ and 1δ tryptophan molecules would show only one pronounced feature close in images.According to these considerations,molecular models of the trimer and tetramer are presented in Figs.2c and e,where intermolecular interactions are maximized mainly through O...H...N hydrogen bonds.We suggest these molecules are at the zwitterionic state which enhances the interaction between tryptophan molecules.When arriving at the cold Cu(111) substrate, the neutral tryptophan molecules might change to the zwitterionic state.Both trimer and tetramer contain configurations 1 and 2 and orientations λ and δ.The proposed molecular models are suggested after considering all possible combinations of configurations and orientations.Besides,tryptophan dimer chains and hexamers are also observed and will be discussed below in detail.
The high-resolution STM image of a dimer chain is amino acid molecules have been demonstrated by Yitamben et al.when Lmethionine molecules were adsorbed on Cu(111) [25].A clear difference is that one had an unambiguous bright protrusion at the molecular end.Interestingly,each dimer in the chain contains two different molecules.The configuration 1λ is consistent with the molecular STM images in the chain(Fig.3a).A stable chain can not be obtained by using only the zwitterionic chemical state for tryptophan molecules to explain the supramolecular structures.It gets stabilized when assuming that the molecule with the dotted end is at the neutral state.The models of twelve molecules are superimposed on the right part of the STM image shown in Fig.3a,which is magnified in Fig.3b to get a clear picture about intermolecular interaction.Two chains are connected through O...H...O hydrogen bonds between carboxyl and carboxylate groups (Fig.3c).Along the chain direction, tryptophan molecules interact with each other through O...H...N hydrogen bonds(Fig.3d).
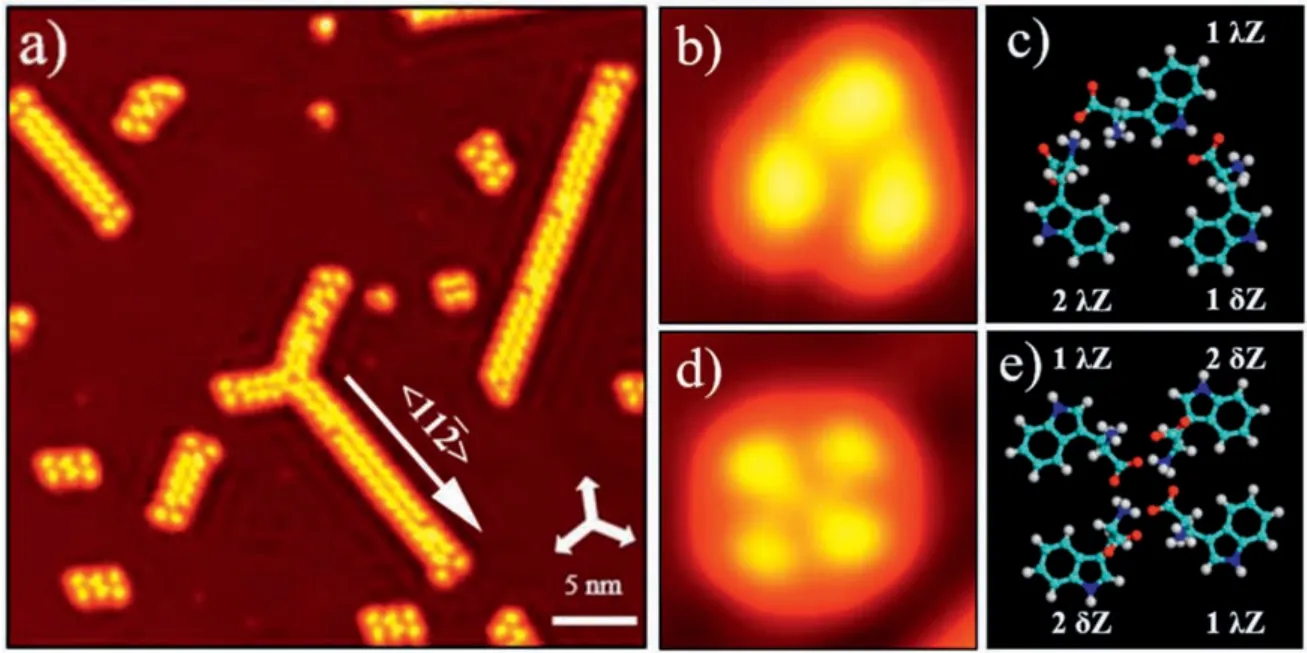
Fig.2.Self-assembly for 0.1 ML coverage of the amino acid L-tryptophan on Cu(111)surface at low temperature.(a)STM image(V=0.5 V,I=40 pA)of L-tryptophan on Cu(111)with the coverage of 0.1 ML after being annealed at 120 K.(b,c) High-resolution STM image of a trimer (3×3 nm2, V=1 V, I=20 pA) and its molecular model.(d,e) Highresolution STM image of a tetramer(3.6×3.6 nm2,V=0.1 V,I=20 pA)and its molecular model.The three white arrows at the right-down corner in(a)indicate the substrate<1 1 0>direction.
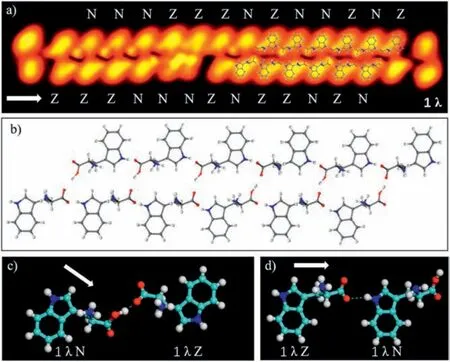
Fig.3.(a) Close-up STM image (14.3×3.6 nm2, V = 50 mV, I=20 pA) of a double chain with twelve molecular models superimposed on the image.We label the zwitterionic state as Z and the anionic state as N.(b) Detailed molecular models shown in(a).(c)A molecular dimer containing two molecules from different chains where the main interaction is O...H...O hydrogen bond.(d) A molecular dimer containing two molecules from a same chain where the main interaction is O...H...N hydrogen bond.In molecular models,hydrogen bonds are indicated by dashed lines.
When annealing at room temperature, tryptophan hexamers dominate the surface (Fig.4a).The hexamers adsorb along the <1 1> orientations (Fig.4b), same as the dimer chains, which indicates a strong interaction between tryptophan and Cu(111).All hexamers have the same molecular arrangement, reflecting unneglected intermolecular interactions in hexamers.Additional chirality can be generated through surface adsorption of tryptophan molecules.Because of the certain interaction between the chiral tryptophan and Cu(111), all hexamers are in the same chirality.A high-resolution STM image of a hexamer and its molecular models are displayed in Figs.4c and d, respectively.As shown in the model, O...H...N hydrogen bonds between tryptophan molecules are the main driving force for the formation of hexamers.The detailed statistical diagram of various tryptophan supramolecular structures is shown in Fig.4e.It indicates that 88.4% of molecules form hexamers.The structural evolution of tryptophan from double chains to hexamers originates from a subtle balance of molecule-substrate and intermolecular interactions.We suggest that all molecules in hexamers are at the anionic state for the following two reasons.On the one hand,other similar amino acids such as glycine[1-3],alanine[5,6]and serine[8]are all proved to be the anionic form when adsorbed on Cu surfaces at room temperature.On the other hand, tryptophan molecules can diffuse freely to some extent on Cu(111) at low coverages at room temperature to reach the energetically stable anionic state via deprotonation.It should be noted that the hexamer is surrounded by positively charged hydrogen atoms,which prohibits the aggregation of hexamers.In addition,the longrange intermolecular interactions induced by the interference of surface state electrons further stabilize the dispersed hexamers.

Fig.4.(a,b) Large-scale (180×180 nm2, V=1 V, I=20 pA) and high-resolution(14.4×14.4 nm2, V = 5 mV, I=200 pA) STM images of homochiral L-tryptophan hexamers on Cu(111) at low coverage (~0.2 ML) after being annealed at room temperature.The three white arrows in the down-right corners of STM images(a,b)indicate the substrate <1 1 0>direction.(c,d)Magnified STM image(3.1×2.9 nm2,V=1 V, I=20 pA) and its molecular model.(e) Statistical diagram of various Ltryptophan supramolecular structures on Cu(111) at low coverage after being annealed at room temperature.
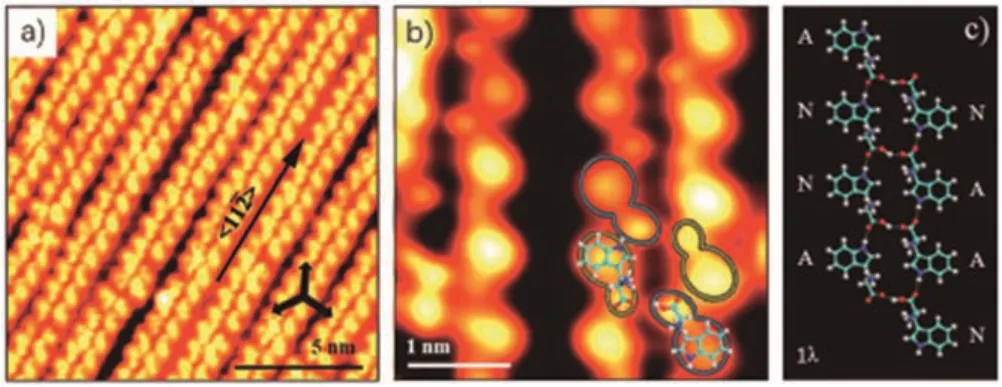
Fig.5.(a)STM image(14.4×14.4 nm2,V=1 V,I=20 pA)of L-tryptophan on Cu(111)with the coverage of 1 ML after being annealed at room temperature.Three black arrows indicate the substrate <1 1 0>direction.(b) High-resolution STM image measured at constant-height mode (3.6×3.6 nm2, V = 5 mV, I=60 pA) revealing two species of L-tryptophan.The blue and yellow calabash-shape curves mark the neutral (labeled as N) and anionic (labeled as A) molecules, respectively.(c)Molecular model for four dimers shown in the previous STM image (b).
Interestingly, a second type of dimer chain (Fig.5a) is experimentally observed when the coverage is increased to 1 ML and the sample is annealed at room temperature.The highresolution STM image measured at constant-height mode(Fig.5b)reveals that the dimer chain is also composed of two species of molecules, which are marked by blue and yellow calabash-shape curves.According to their appearance in STM images, we suggest all molecules in the chains adopt the 1λ configuration.One kind of species should be at the anionic state rather than zwitterionic form because of the relatively high annealing temperature.To maximize intermolecular interactions, another kind of species should be at the neutral state.After comparing molecular heights in STM images, the bright and dark tryptophan molecules are at the anionic and neutral states, respectively.The molecular model of four dimers in the chain is shown in Fig.5c.The existence of neutral molecules at room temperature might be caused by the competition between intermolecular interaction and molecule-substrate interaction.The stronger molecular interaction reduces the impact of the substrate and makes some molecules in the neutral form.In addition, tryptophan molecules can not diffuse freely at this high coverage, which decreases the possibility of the dehydrogenation process.
In summary, we investigate the self-assembly of L-tryptophan molecules on the Cu(111) surface by an ultrahigh vacuum lowtemperature scanning tunneling microscope.At low coverages,tryptophan molecules form trimers, tetramers, hexamers, and chains after annealing the sample at around 120 K.Further increasing the annealing temperature to 300 K leads to a clear supramolecular structure conversion to hexamers.They are dispersed on the substrate because of the repulsion from molecular peripheral hydrogen atoms and the long-range intermolecular interactions induced by the interference of surface state electrons.At high coverages, a different type of chain is observed when the sample is annealed at room temperature because of a subtle balance between intermolecular and molecule-substrate interactions.According to high-resolution STM images with the resolution of sub-molecular level,we suggest that the L-tryptophan molecules are present in neutral, zwitterionic or anionic states in these structures.
Acknowledgments
This work is supported by the Ministry of Science and Technology( Nos.2018YFA0306003, 2017YFA0205003) and National Natural Science Foundation of China ( No.21972002).DFT calculations are carried out on TianHe-1A at National Supercomputer Center in Tianjin and supported by High-performance Computing Platform of Peking University.
杂志排行
Chinese Chemical Letters的其它文章
- A roadway of exploring polymer science, a lifetime of nurturing polymer scientists
- A personal journey on using polymerization in aqueous dispersed media to synthesize polymers with branched structures
- Amphiphilic block copolymers directed synthesis of mesoporous nickel-based oxides with bimodal mesopores and nanocrystal-assembled walls
- Synthesis of magnetic polyphosphazene-Ag composite particles as surface enhanced Raman spectroscopy substrates for the detection of melamine
- Photothermal performance of MFe2O4 nanoparticles
- Enhanced electrochemical performance and mechanism study of AgLi1/3Sn2/3O2 for lithium storage
