Cu-DNAzyme facilitates highly sensitive immunoassay
2019-09-24ZhiShnMingshengLyuDennisCurryDviOkleyKenOkesXuZhng
Zhi Shn,Mingsheng Lyu,Dennis Curry,Dvi Okley,Ken Okes,Xu Zhng,e,*
a Verschuren Centre for Sustainability in Energy & the Environment, Cape Breton University, Sydney, Nova Scotia, B1P 6L2, Canada
b College of Life Science, Sichuan Agriculture University, Yaan 625014, China
c Jiangsu Key Laboratory of Marine Bioresources and Environment, Huaihai Institute of Technology, Lianyungang 222005, China
d Department of Biology, Cape Breton University, Sydney, Nova Scotia, B1P 6L2, Canada
e Department of Chemistry, Cape Breton University, Sydney, Nova Scotia, B1P 6L2, Canada
Keywords:Cu-DNAzyme Immunoassay ELISA CuO Nanoparticles
ABSTRACT Fast and sensitive antigen detection is important in biomedical research and development.Despite,being invented 48 years ago, the enzyme-linked immunosorbent assay (ELISA) remains one of the most successful and widely employed bioanalytical techniques in research and clinical diagnostics due to its reliability and simplistic design.Recently,nanotechnology has offered efficient signal reporting.In spite of some improvements in these systems, there are typically increased material costs involved and the need for expensive equipment or complicated chemical processes, thus negating any possible benefits over ELISA. Herein, we communicate a simple Cu-DNAzyme system for signal transduction of a CuO nanoparticle-labeled immunoassay. The reported immunoassay amplifies signal generation similar to traditional ELISA and is fast, simple, cost-effective, and sensitive, holding promise for biomedical applications and point-of-care testing.
Due to the heightened interest in point-of-care(POC)diagnostic systems, rapid, cost-effective and easy to use biomolecule detection methods remain highly desirable [1-3]. The first description of enzyme-linked immunosorbent assay (ELISA) in 1971 by Egvall and Perlemen marked a watershed moment in antigen detection and biomedical diagnostics [4]. The simplistic nature of the assay’s design, relying only on antigen/antibody immobilization, recognition and signal transduction, has since allowed for the sensitive and robust detection of a host of antigen molecules. Following antibody labelling using enzymes such as horseradish peroxidise (HRP), oxidation of a chromogenic substrate molecule(e.g.,3,3′,5,5′-tetramethylbenzidine,TMB)leads to sensitive colorimetric detection. Since its initial description 45 years ago,little has changed with respect to the basic design of the immunoassay.Recent innovations in nanotechnology and artificial enzyme development have allowed for increased sensitivity based on novel signal transduction mechanisms [5-17]. In these immunoassays,nanoparticles(NPs)serve as either more sensitive reporters (e.g., gold NPs offer sensitive optical readouts while fluorescent silicon NPs and quantum dots serve as attractive fluorescence labels) or as an efficient carrier of the reporter molecules. Furthermore, new signal amplification mechanisms have been adopted such as rolling circle amplification (RCA) and those involving metalloDNAzymes to improve assay sensitivity[18,19]. Nevertheless, many of the reported systems require expensive or even toxic chemicals and complicated materials synthesis, prolonged assay time, or expensive instrumental analysis, making these systems unsuitable for POC practice in remote and low-resource community settings [18,20].
Herein, we communicate simple but highly sensitive Cu-DNAzyme-based immunoassay readout (Scheme 1), where costeffective CuO NPs were used to label the detection antibody in the immunoassay. After the antigen recognition reaction by CuO NP-labeled antibody(CuO@rabbit anti human IgG),copper ions are released by a 5-min acid-treatment [20-22]. The released copper ions then interact with a single-stranded homopolymer polyguanine sequence (i.e., G20) to form an efficient Cu-DNA-based peroxidise mimic (herein referred as to Cu-DNAzyme), which serves as a homogeneous catalyst to accelerate the oxidation of TMB,generating a bluish signal for sensitive colorimetric detection[23-25].The proposed DNAzyme provides substantial advantages over traditional ELISA enzymes(i.e.HRP-based)such as decreased cost, increased chemical stability and shelf-life, and easy transportation and storage.The Cu-DNAzyme immunoassay differs from traditional ELISAs with respect to its rapid and simple antibody-labelling protocol (herein involving only simple mixing of the detection antibody with CuO NPs in pure water followed by a brief centrifugation step to remove free antibody), generating stable CuO@antibody conjugates(within 12 h),as shown in Fig.S1(Supporting information), and highly efficient colorimetric readout. The homogeneous DNAzyme facilitates signal generation in just 5 min; in contrast, in the traditional ELISA, HRP must be immobilized on the plate surface via conjugated antibody,catalyzing signal generation in a more time-consuming (e.g.,30-60 min) heterogeneous manner. Without the need for expensive enzyme molecules,tedious antibody-labeling procedures,and advanced instrument detection, the proposed immunoassay system is time- and cost-effective and importantly, retains the trademark strengths of ELISA including ultra-high sensitivity for instrument quantification and suitability for naked-eye detection with its bluish oxidized product.
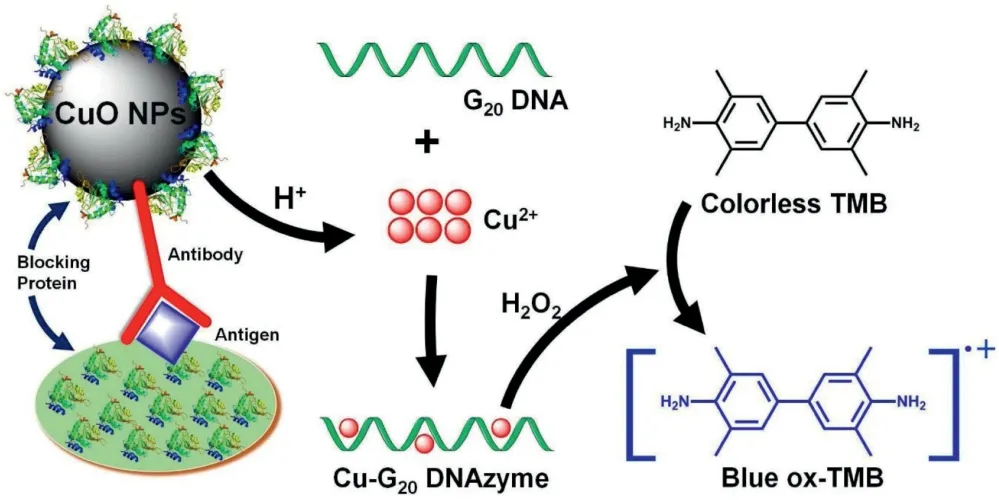
Scheme 1. Cu-polyG DNAzyme direct immunoassay for highly sensitive detection of antigen molecules.
The readout mechanism is based on the high HRP-like activity of the Cu-G20complex,which forms a simple butefficient DNAzyme.In this ELISA,G20sequencewaschosen due to its cost-effectiveness and capabilityto generate signal with the best sensitivity,although other G-rich sequences (Fig. S2a in Supporting information) can also be used for the colorimetric assay as systematically demonstrated in Y.Fu’s work[23-25].As shown in Fig.S2(Supporting information),the Cu-G20DNAzyme catalyzes TMB oxidation by H2O2,generating a bluish product in a quantitative manner,of which the color intensity strongly correlates to the concentration of the Cu-DNAzyme in the sample solution.Similar to proteinaceous enzymes such as HRP,the catalytic efficiency of the DNAzyme is pH dependent and catalytically optimal at pH 4(Fig.S3 in Supporting information)[26,27].The Cu-DNAzyme offers fast reaction kinetics (5-10 min) (Fig. S4 in Supporting information),ultrahigh sensitivity(detection limit:2.68nmol/L),a wide dynamic range(2.68 nmol/L-80 μmol/L)(Fig.S5 in Supporting information), high selectivity and low interference by most co-existing metal ions(note:Fe2+can interfere with the Cu2+detection as shown in Fig. S6 (Supporting information), but in immunoassay systems, free Fe2+concentrations are typically negligible).
The effect of bovine serum albumin (BSA), casein, or protein matrix (fetal bovine serum: FBS) on background noise and signal intensity during colorimetric assays for Cu ions was evaluated since these proteins are often involved in immunoassays either in the sample or as blocking agents for microplates[28-30].Neither BSA,casein,nor FBS,increased background noise in the absence of copper(Fig.S7 in Supporting information).To assess the impact of these proteins on signal intensity,copper detection was carried out with proteins either suspended in solution(Fig.S8a in Supporting information)or adsorbed to plate well surfaces(Figs.S8b and c in Supporting information). Casein, even when suspended in solution, exerted little effect on signal intensity and is therefore most suitable for blocking purposes, while suspended BSA affected signal intensity at low copper concentrations. However, when these proteins were applied in a pre-coating protocol as plate blocking agents, neither BSA nor FBS affected signal intensity,demonstrating the Cu-DNAzyme-based colorimetric assay is wellsuited for immunoassay applications utilizing FBS,BSA or casein as they may be used without negatively impacting background noise or signal intensity.
Given the sensitive copper detection capabilities of the simple Cu-G20DNAzyme method, we utilized the platform for immunoassay readout. Signal transduction began by labelling antibodies with CuO NPs via simple mixing and centrifugation.The feasibility of the immunoassay was demonstrated using human IgG as a model antigen(i.e.,the analyte)and rabbit anti-human-IgG as the targeting antibody. Upon adsorption of the target antigen in 96-well plates, several reagent combinations were used to systematically test selectivity of the immunoassay. Fig. 1 shows the chromogenic test result of reagent combinations used in each experimental well.Well 1 represented the proposed immunoassay system,including both the antigen and antibody-labelled CuO NPs and showed a positive test result,signifying effective detection of the target antigen. Well 2, without the target antigen or blocking proteins, showed a false positive result, albeit with less color intensity than well 1,where color development is attributed to the non-specific binding of the antibody-labelled CuO NPs to the well surface due to the absence of casein as a blocking protein. In contrast, wells 9 and 10, which lacked the target antigen but included the blocking protein (control for well 2), produced a negative test result,demonstrating the importance of the blocking protein to ensure assay accuracy and selectivity. Wells 3-8 varied in reagent composition, serving as controls investigating the potential for unanticipated false positive results and the effects of individual components on signal amplification. The results show that wells lacking antibody-labelled CuO NP (wells 3-8) did not result in color development. Collectively, this experiment result shows the feasibility of the proposed assay in antigen detection and demonstrates the need for effective protein blocking,similar to established immunoassay systems [28,31].
The performance of the Cu-G20system was then compared to the commercially-available HRP-based ELISA immunoassay protocol[32].Fig.2 demonstrates the Cu-G20immunoassay is effective and more sensitive (the detection limit and quantification limit were 0.1 ng/mL and 3.4 ng/mL, respectively) than the traditional HRP-based ELISA (detection limit: 0.3 ng/mL). Moreover, the Cu-G20DNAzyme retains the additional advantage of a very simple antibody labelling procedure, use of a less-expensive (and more stable)enzyme mimic,and faster signal generation.To evaluate the potential of the Cu-G20system for in vitro clinical diagnostic applications, we prepared artificial human serum samples by spiking human IgG(the analyte)into FBS(the mimic of real sample matrix) at concentrations of 1, 0.1 and 0.01 mg/mL (10, 100 and 1000 time dilutions of human blood,respectively)[33].The results clearly demonstrate the feasibility of the proposed system for in vitro antigen detection in the context of clinical diagnostics(Fig.3),even roughly quantifiable(within orders of magnitude)by the unaided eye, a critical attribute for POC applications.
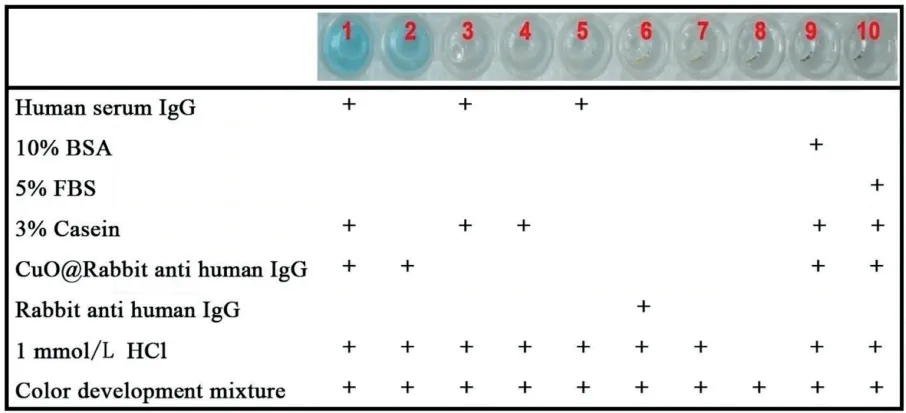
Fig.1. Selectivity of the proposed method for specific detection of human IgG.The reagents added to each well are indicated below the images, which were taken 5 min after adding color development mixture to each well. Color development mixture: 1μmol/L G20, 0.5 mmol/L TMB, 500 mmol/L H2O2 in pH 4.0 MES buffer.
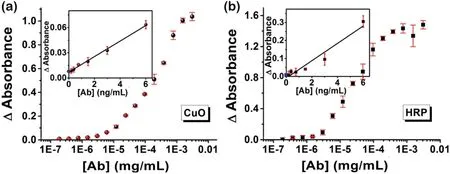
Fig.2. Comparison between(a)the proposed colorimetric Cu-DNAzyme immunoassay and(b)the standard HRP-based ELISA for human IgG.Inset:the signals(light absorbance)vs.low concentrations of analyte(human IgG),where the signals on yaxis (when [Ab]=0 ng/mL) stand for the background noise level.

Fig. 3. Detection of human IgG in serum samples.
In conclusion, we have described a Cu-DNAzyme system for immunoassay signal transduction. Signal amplification in the system is similar to that of traditional ELISA platforms, whereby the DNAzyme-mediated generation of reactive species leads to TMB oxidation, resulting in sensitive colorimetric readout.Compared to other metal oxide nanoparticle-based immunoassays(Table S1 in Supporting information),this system provides a fast (e.g., the antibody conjugation time is only 10 min, much quicker than any other methods), simple, cost-effective method for high sensitivity antigen detection, and may be of particular interest to those working in medical diagnostics and point-of-care testing.
Acknowledgments
This work was supported by Beatrice Hunter Cancer Research Institute, the ACOA AIF program, CBU RISE program, Nova Scotia Lands,Innovacorp ESCF Phase I grant,the New Frontiers in Research Fund-Exploration,Canada Foundation of Innovation John R.Evans Leaders Fund, the Priority Academic Program Development of Jiangsu Higher Education Institutions,and NSERC Discovery Grants.
Appendix A. Supplementary data
Supplementarymaterialrelatedtothisarticlecanbefound,inthe online version,at doi:https://doi.org/10.1016/j.cclet.2019.05.037.
杂志排行
Chinese Chemical Letters的其它文章
- Etching of gold nanorods: The effects of diameter on analytical performances
- Target-triggered inhibiting oxidase-mimicking activity of platinum nanoparticles for ultrasensitive colorimetric detection of silver ion
- Gold nanoparticles as dehydrogenase mimicking nanozymes for estradiol degradation
- SciFinder-guided rational design of fluorescent carbon dots for ratiometric monitoring intracellular pH fluctuations under heat shock
- Highly selective electrochemical method for the detection of serotonin at carbon fiber microelectrode modified with gold nanoflowers and overoxidized polypyrrole
- Facile and efficient fabrication of g-C3N4 quantum dots for fluorescent analysis of trace copper(II) in environmental samples
