Clinical significance of MET gene amplification in metastatic or locally advanced gastric cancer treated with first-line fluoropyrimidine and platinum combination chemotherapy
2019-09-10SeyoungSeoMinHeeRyuBaekYeolRyooYangsoonParkYoungSooParkYoungSoonNaChaeWonLeeJuKyungLeeYoonKooKang
Seyoung Seo, Min-Hee Ryu, Baek-Yeol Ryoo, Yangsoon Park, Young Soo Park, Young-Soon Na,Chae-Won Lee, Ju-Kyung Lee, Yoon-Koo Kang
1Departments of Oncology; 2Department of Pathology; 3Asan Institute for Life Science, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, Republic of Korea
Abstract Objective: To investigate the clinical significance of MET gene amplification in patients with gastric cancer in the palliative setting.Methods: MET amplification was assessed using fluorescence in situ hybridization (FISH) in 50 patients and quantitative polymerase chain reaction (qPCR) in 326 patients; 259 patients treated with first-line fluoropyrimidine and platinum were included for survival analysis.Results: The results of FISH and qPCR indicated that the c-MET/CEP7 ratio was correlated with gene copy number. The optimal cutoff value for the copy number using qPCR to detect MET gene amplification with FISH was 5 (κ=0.778, P<0.001). Twenty-one out of 326 patients (6.4%) were identified as MET amplification with a copy number of >5 detected by qPCR. MET-amplified gastric cancer was associated with an Eastern Cooperative Oncology Group (ECOG) performance status (PS) score of ≥2 (33.3% vs. 10.5% P=0.007), peritoneal metastasis(76.2% vs. 46.2%, P=0.008), and elevated bilirubin levels (28.6% vs. 7.3%, P=0.006). The median overall survival(OS) and progression-free survival (PFS) were 11.9 and 5.6 months, respectively. MET-amplified gastric cancer was not associated with survival outcomes [hazard ratio (HR)=0.68, 95% confidence interval (95% CI): 0.35-1.32,P=0.254 for PFS; HR=0.68, 95% CI: 0.35-1.32, P=0.251 for OS].Conclusions: qPCR can be used to detect MET gene amplification. MET amplification was not a predictor of poor prognosis in patients with metastatic or unresectable gastric cancer.
Keywords: MET; amplification; advanced gastric cancer; prognosis; quantitative real-time polymerase chain reaction
Introduction
Despite improvements in outcomes with targeted agents,including trastuzumab (1), ramucirumab (2) and apatinib(3), the prognosis of unresectable or metastatic gastric cancer remains unfavourable. There are unmet needs to discover novel treatments for advanced gastric cancer.
Dysregulation of c-MET signaling pathway has been implicated not only in gastric cancer (4), but also in other malignancies, including breast, lung, pharynx, colorectal,and cervical cancers (5-9). The aberrant c-MET signaling pathway including gene mutation, gene amplification,overexpression of the ligand and/or receptor, autocrine signaling, and paracrine signaling has been indicated as a potential mechanism in carcinogenesis (10). The activation of c-MET signaling pathway in gastric cancer has been associated mainly with gene amplification (11-13), whereas gain of function mutations in MET are rare (14). MET proto-oncogene amplification activates the MET/hepatocyte growth factor pathway to promote cell proliferation, anti-apoptotic activities, cell detachment,migration, and invasion for metastasis (15,16).
Fluorescence in situ hybridization (FISH) is the standard method to detect gene amplification. Because of the high cost and long turnaround time for obtaining FISH results,a real-time quantitative polymerase chain reaction (qPCR)-based gene copy number assay has been considered as an alternative method for gene amplification. However, the cutoff value of proper copy number for predicting MET gene amplification has not yet been established. The frequencies of MET gene copy number gain in gastric cancer ranged from 1.5% to 21.2% depending on the cutoff points (17-19). In addition, the concordance between MET gene amplification using qPCR and FISH is controversial (17-20).
Little is known about the clinicopathologic features and prognosis of MET-amplified gastric cancer, although studies suggested worse survival outcomes for METamplified gastric cancer patients based on resected tissue obtained during curative surgery (11,17,18,20). Based on these findings, there are some limitations for patients with metastatic or unresectable gastric cancer who are indicated for palliative chemotherapy.
In this study, we investigated the efficacy of qPCR to screen MET gene amplification and determined the clinicopathologic features and prognosis of MET gene copy number gain in patients with locally advanced unresectable or metastatic gastric cancer.
Materials and methods
Study population and data collection
Two types of registries were used in this study. The first one was a retrospective registry consisting of 552 patients with locally advanced or metastatic gastric cancer who were treated with a first-line fluoropyrimidine and platinum (FP)regimen between June 2006 and June 2011. After histological review, 193 cases with ≥70% tumor cells in the formalin-fixed paraffin-embedded (FFPE) tissues were selected. The second one was a prospective registry that consisted of 815 patients with locally advanced or metastatic gastric cancer between September 2012 and December 2014, who were not categorized according to chemotherapy regimen. All collected FFPE tissues were examined to define the tumor cell proportion and gene copy number using qPCR. There were 133 patients with gene amplification based on qPCR results in patients with≥70% tumor proportion. Among them, only 66 patients received a first-line FP regimen. In addition, we randomly selected 50 patient samples from a prospective gastric cancer registry to assess MET gene amplification using both FISH and qPCR. Tumor microdissections were conducted by a pathologist (Y Park), when the sample did not have sufficient tumor cells (<70%) before conducting FISH and qPCR.
This study adhered to the guidelines established by the Declaration of Helsinki, and was approved by the Institutional Review Board at Asan Medical Center.
FISH
For FISH, 2-μm sections from each paraffin block were prepared. Deparaffinization, pretreatment and protease digestion procedures were performed following an established protocol using a D7S522 probe and CEP7 purchased from Abbott Vysis (Des Plaines, IL, USA).Probes were hybridized at 37 °C for 14-18 h. After hybridization, slides were washed in 2× saline-sodium citrate/0.3% NP-40 at 72 °C for 5 min, air-dried, and counterstained with 4',6-diamidino-2-phenylin-dole(DAPI). The slides were examined under a fluorescence microscope equipped with Spectrum Texas Red,fluorescein isothiocyanate, and DAPI filters. The slides were stored at -20 °C until examination. The cMET/CEP7 ratio was established after counting at least 40 tumor cells. A cMET/CEP7 ratio of ≥2 and ≥10%tumor cells with a MET gene copy number >4 were considered as MET gene amplification.
Real-time qPCR-based determination of gene copy number
Genomic DNA was extracted from biopsies or surgical FFPE tissues using a QIAamp DNA FFPE Tissue kit or QIAamp DNA Mini kit (Qiagen, Hilden, Germany). The DNA concentration was measured using a NanoDrop2000 spectrophotometer (Thermo Scientific, Waltham, MA,USA). Predesigned Applied Biosystems TaqMan copy number assays were performed (Thermo Scientific,Waltham, MA, USA) to determine the gene copy number of MET. A total volume of 10 μL of the Master mix contained 10 ng of genomic DNA, 5 μL of the TaqMan genotyping Master mix, and each primer for real-time PCR. The primer ID was Hs02884964_cn. The telomerase reverse transcriptase gene and human genomic DNA(Takara) were used as the internal reference for the copy number and the normal control, respectively. The thermal cycling conditions were 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. The results were analyzed using the ABI PRISM 7900HT Sequence Detection System (Thermo Scientific, Waltham, MA).
Statistical analysis
The Spearman's rank correlation coefficient was used to assess the correlation between FISH and qPCR data. A receiver operating characteristic (ROC) curve was plotted and the area under curve (AUC) was estimated to set the cutoff value of the highest sensitivity and specificity by calculating the κ values using concordance evaluation.
Progression-free survival (PFS) was defined as the time between the start of FP chemotherapy to tumor progression or death by any cause. Overall survival (OS)was calculated from the initiation date of first-line FP to death by any cause. Data were censored if patients were free from progression or alive at the last follow-up.Categorical variables were evaluated using the Chi-square test or Fisher's exact test, as appropriate. The Kaplan-Meier method was used to estimate PFS and OS. Survival curves were compared using a log-rank test, according to MET amplification. A Cox proportional hazard model was used to estimate hazard ratio (HR) for survival outcomes.
All statistical analyses were performed using the IBM SPSS Statistics (Version 21; IBM Corp., NewYork, USA)for the Social Sciences and statistical software package R version 3.0.2 (http://www.r-project.org/). All tests were two-sided with 5% defined as the level of significance.
Results
Correlation of copy numbers evaluated using qPCR and FISH
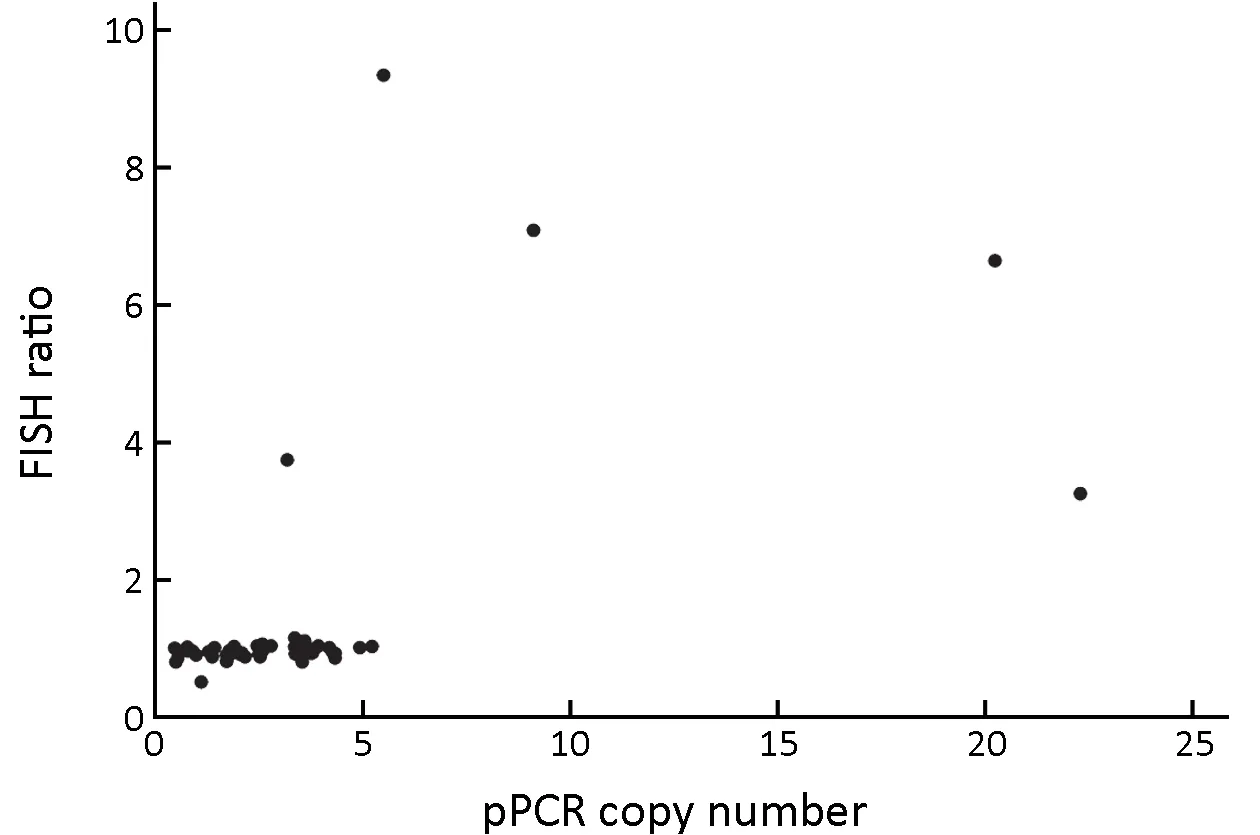
Figure 1 Correlation between MET gene copy numbers and c-MET/CEP7 ratios in 50 metastatic gastric cancer patients. FISH,fluorescence in situ hybridization; qPCR, quantitative polymerase chain reaction.
The analysis of FFPE specimens with both qPCR and MET FISH revealed a good correlation between the c-MET/CEP7 ratios and the copy numbers with a value of 0.443 (P=0.002) based on Spearman's rank correlation coefficient. Five cases (10%) showed MET gene amplification with a c-MET/CEP7 ratio of greater than 2(Figure 1). One patient without MET amplification had a c-MET/CEP7 ratio of 0.82 and a copy number of 2.98(Figure 2A). Among the 5 patients with MET gene amplification, the range of cMET/CEP7 ratio was from 3.75 to 9.35 and the copy numbers ranged from 7.18 to 9.03 (Figure 2B,C). To determine the cutoff point of copy numbers to assess MET gene amplification, we plotted a ROC curve with AUC 0.953 (Supplementary Figure S1).The value of the highest sensitivity and specificity was identified at 5.22 copy numbers. We calculated the κ values at the cutoff point of various copy numbers. The κ values for 3, 4, 5, 6, and 7 copy number were 0.248 (P=0.008),0.462 (P<0.001), 0.778 (P<0.001), 0.730 (P<0.001), and 0.730 (P<0.001), respectively.
Patient characteristics
A total of 326 samples (64 samples from surgical resection and 262 samples from biopsy) were suitable for assessing the association between MET amplification and clinicopathologic factors. Among them, 259 patients treated with first-line FP were used for the survival analysis.
The median age of patients was 58 (range, 23-85) years old. Among all patients, 65.9% of patients had initially metastatic disease, and others presented with recurrence and locally advanced unresectable disease. At the time of diagnosis, 287 (88.0%) patients had an Eastern Cooperative Oncology Group (ECOG) performance status (PS) score of 0-1 (Table 1). The distant lymph nodes and peritoneum were the most common metastatic sites, and more than half of the patients (62.3%) presented with poorly differentiated histology. 122 (37.4%) patients did human epidermal growth factor receptor 2 (HER2) immunohistochemistry(IHC) and/or silver in situ hybridization, and 103 (31.6%)patients were HER2 negative. Nineteen (5.8%) HER2+patients were determined based on HER2 positivity with IHC 3+ or IHC 2+ gene amplification by in situ hybridization.
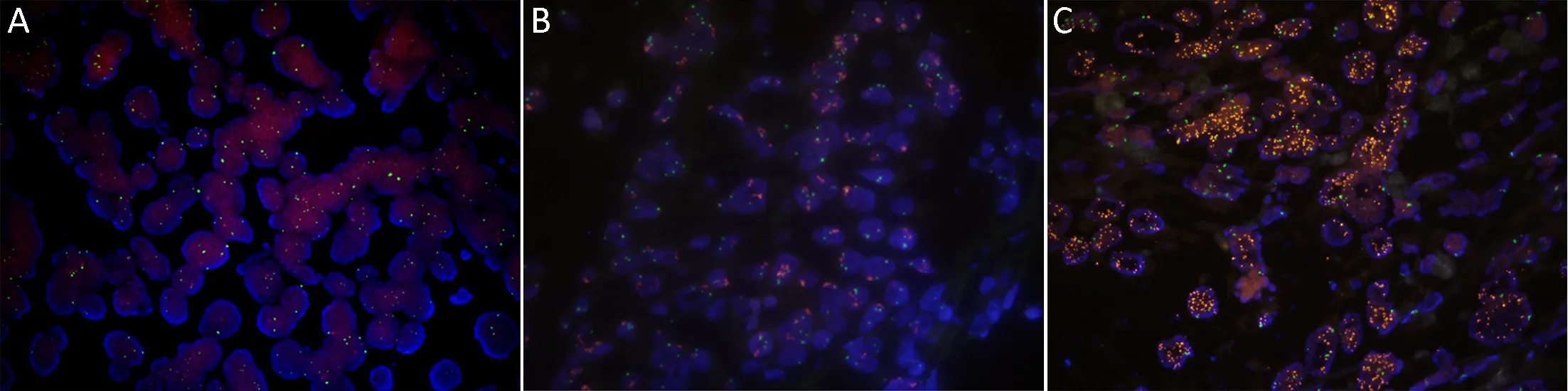
Figure 2 MET gene amplification was evaluated using fluorescence in situ hybridization showing representative image of non-amplification(A), low-level amplification (B), and high-level amplification (C).
The median copy number of MET using qPCR was 1.69 with a range of 0.18—206.30. When applying the optimal cutoff point of 5 copy numbers based on qPCR results to detect MET amplification using FISH, the frequency of MET amplification was 6.4% (n=21).
Association of MET amplification with clinicopathologic features
Clinical characteristics were compared between patients with and without MET amplification using qPCR with a cutoff point of >5 copy numbers. The MET amplification group had an ECOG PS of ≥2 (33.3% vs. 10.5%, P=0.007),lower albumin level (52.4% vs. 31.6%, P=0.051), and elevated total bilirubin (28.6% vs. 7.4%, P=0.006)compared with those of the non-amplification group.When we applied our previously developed prognostic model for metastatic or recurrent gastric cancer (21), MET amplification was associated with the poor prognostic group (47.6% vs. 14.2%, P=0.001) (Table 2). This prognostic model divided patients into three risk groups according to the sum of scores (good; 0-1, moderate; 2-3 and poor; ≥4) which was based on 8 clinical features;ECOG PS (score 2), no gastrectomy history, peritoneal metastasis, bone metastasis (score 2), lung metastasis,elevated alkaline phosphatase, decreased albumin level and increased bilirubin level.
Association of MET amplification with survival outcome
Among 259 patients treated with an FP regimen, 171 patients presented with measurable lesions, and 81 (47.4%)out of 171 patients achieved objective responses. The overall response rate showed no significant difference between the MET amplification group and the nonamplification group [8 of 15 (53.3%) vs. 73 of 156 (46.8%),respectively, P=0.628]. Overall, 94.6% patients died at the time of analysis. With a median follow-up of 12.6 (range,0.7-104.2) months, the median OS and PFS were 11.9[95% confidence interval (95% CI): 10.2-13.6)] months and 5.6 (95% CI: 4.5-6.7) months, respectively.
In the univariate analysis, both PFS and OS were similar between patients with MET-amplified gastric cancer and those with non-amplified gastric cancer (Figure 3). The median OS of the MET-non-amplified and the METamplified groups were 12.6 (95% CI: 10.9-14.4) months and 11.3 (95% CI: 2.2-20.5) months, respectively, and their median PFS were 5.5 (95% CI: 4.4-6.6) months and 5.6 (95% CI: 1.4-9.8) months, respectively. PFS and OS were even worse in patients who had not undergone gastrectomy and had poor PS, Borrmann type IV disease,lung metastasis, bone metastasis, a low albumin level, and an elevated alkaline phosphatase level (Table 3). Using our previous prognostic model, these risk factors showed good discriminative function to predict OS (Table 3).
Since MET amplification was not a significant prognostic factor, multivariate analysis was not conducted with other risk factors. When we conducted the multivariate analysis with the risk groups using our prognostic model and amplification, MET amplification was not significantly associated with either PFS (HR=0.71, 95% CI: 0.39-1.28;P=0.252) or OS (HR=0.67, 95% CI: 0.37-1.20; P=0.189).
Discussion
We assessed MET gene amplification using a qPCR-based gene copy number assay. MET gene amplification was observed in 6.4% patients with metastatic or locally advanced unresectable gastric cancers who received palliative chemotherapy. MET amplification was associated with poor pre-treatment PS, peritoneal metastasis, andelevated bilirubin levels. We found that MET gene amplification was not associated with the prognosis of patients with metastatic or locally advanced unresectable gastric cancers who were treated with palliative FP chemotherapy. To the best of our knowledge, our study is the largest one to investigate the clinical significance of MET amplification in metastatic or locally advanced unresectable gastric cancers.
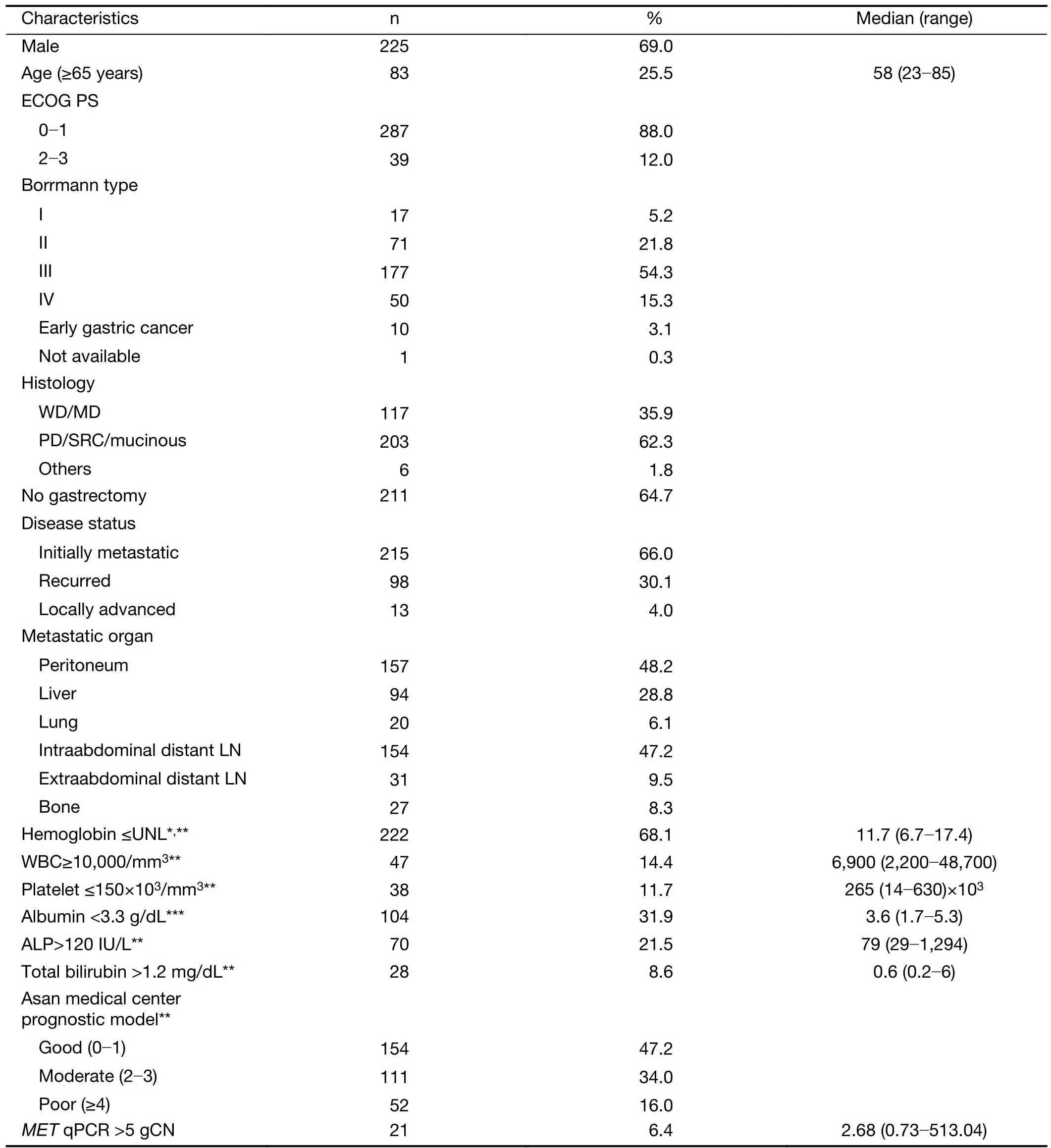
Table 1 Baseline characteristics (N=326)
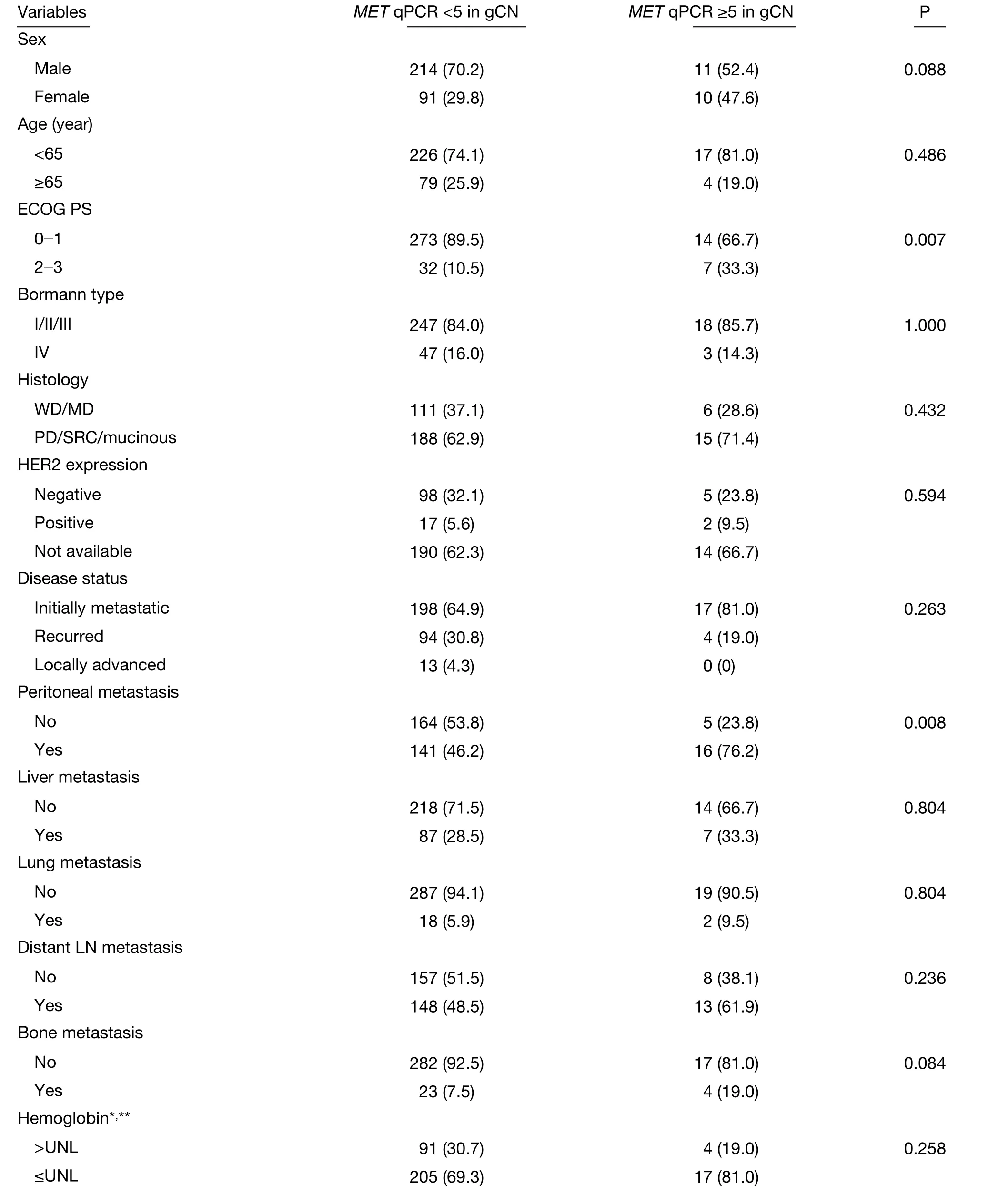
Table 2 Relationships between c-MET amplification and clinicopathologic features (N=326)

Table 2 (continued)
To detect the activation of the c-MET pathway in gastric cancer, various methods have been applied, such as FISH or silver in situ hybridization for gene amplification(12,18,20,22,23), real-time PCR to assess amplification or messenger RNA expression level (17,19,23), and immunohistochemistry for protein level (11,20,23-26).Among them, the ideal surrogate marker to assess the c-MET pathway is inconclusive. Although FISH is the standard method for detecting gene amplification, qPCRbased copy number assays to detect MET gene amplification was explored in our study because FISH is expensive and time-consuming, and it requires technical expertise (27).There were no definite conclusions regarding the appropriate cutoff value for the gene copy number, and the range were 2-5 (17-20). Lee et al. reported that the concordance rate between MET amplification assessed by qPCR and FISH was only 58.1% in 309 tissue samples,when the cutoff value for the copy number was 4 (18).Another study reported a strong correlation between the results of qPCR and silver in situ hybridization with a cutoff value for the copy number >2 in 26 tissue samples (20). Our results suggest that qPCR have similar ability to evaluate MET amplification with FISH with gene copy number of>5. Because the qPCR results depend on the tumor proportion of the samples, we performed tumor microdissection to maintain the tumor cell proportion at more than 70% to minimize the dilution of tumor cells by normal cells, which could explain the good concordance rates between the qPCR and FISH results.
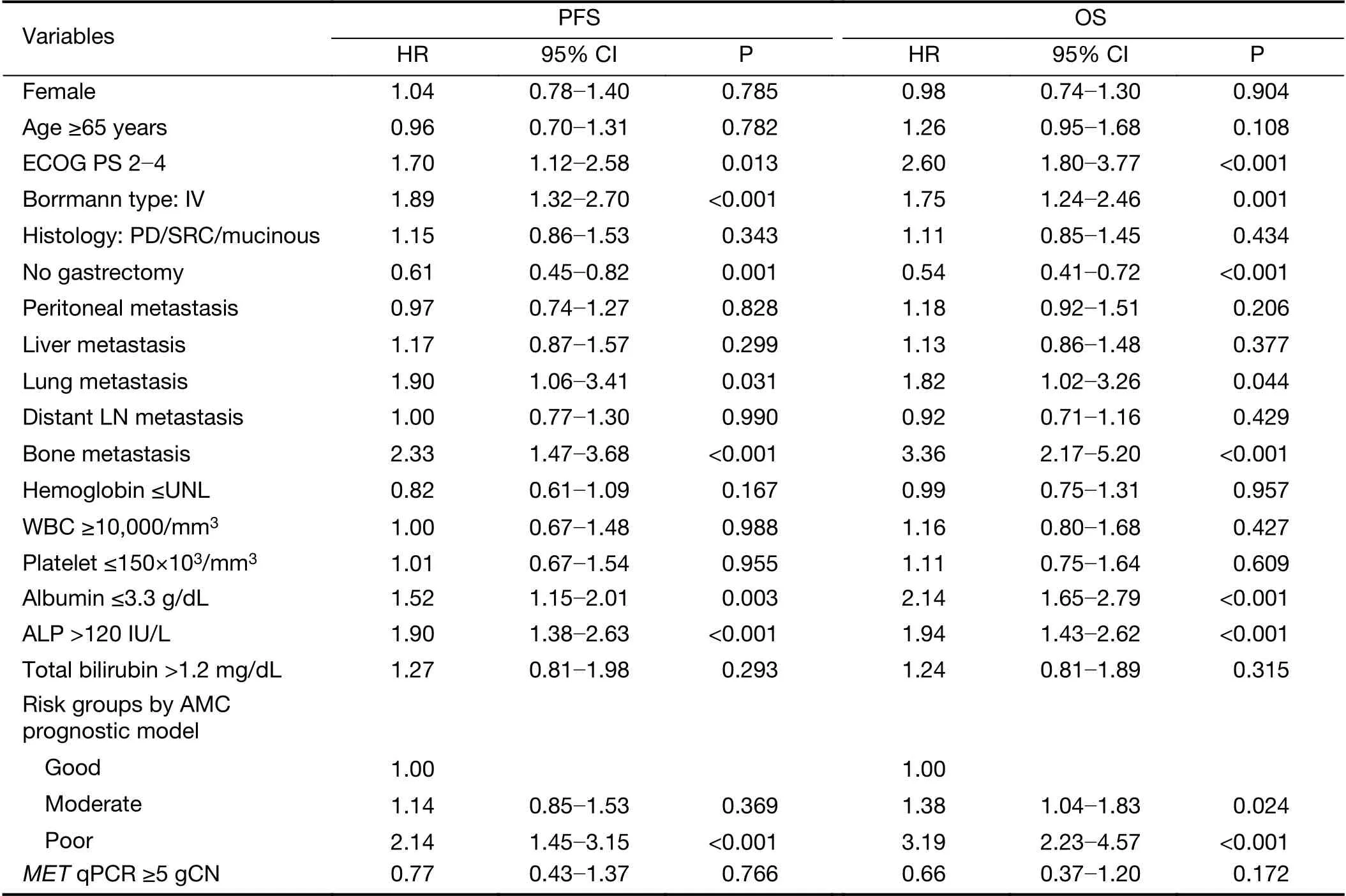
Table 3 Univariate analysis of association of clinicopathologic factors with PFS and OS (n=259)
Previous data from patients with gastric cancer who underwent curative resection showed MET protein overexpression in 22.0%-82.4% cases using IHC(11,24,25), whereas the incidence of MET gene amplification was found to be 1.5%-10.0% using FISH(19,22) and 10.0%-21.2% using qPCR (17,18,23). The inconsistency may be from a poor correlation between high protein expression and MET gene amplification (18,23,24)or from MET gene heterogeneity in surgical tissue (28),although one report demonstrated a good correlation between MET protein expression and gene amplification(20). Various antibodies, different definitions of positivity or cutoff of scoring systems could contribute to the inconsistent results (11,20,23-25). In contrast, the frequency of MET amplification in patients with metastatic or recurrent gastric cancer was 8.3% using FISH (26) and 10.3% using qPCR (29). The frequency of MET amplification detected by qPCR in our study was 6.4%,which is lower than those of previous studies.
Our study revealed that MET gene amplification was associated with poor performance, peritoneal metastasis,and elevated bilirubin levels. One study demonstrated that MET amplification is associated with poor performance and poorly differentiated histology in metastatic gastric cancer(26), and MET protein level is associated with liver metastasis (30). As mentioned above, most of these factors are in turn associated with poor prognosis in metastatic or recurrent gastric cancers (21,31,32). Furthermore, in resectable cases, MET protein level is associated with an advanced disease stage and lymph node metastasis (11,25),and MET gene amplification is also associated with an advanced disease stage (22) and progression to peritoneal metastasis (13). These findings suggest that MET gene amplification might be related to metastatic disease progression in resectable gastric cancer.
Studies have delineated the association between poor OS and MET protein overexpression (11,20) or MET gene amplification (17,18,22) in patients who underwent curative surgery for resectable gastric cancer. In terms of metastatic or unresectable gastric cancer, 2 studies have indicated a poor clinical outcome in MET-amplified gastric cancer patients who received palliative chemotherapy (26,29),whereas biomarker analysis of the RILOMET-1 trial revealed no significant relationship between MET amplification and treatment outcome in a palliative setting(33). Although An et al. used both FISH assay and IHC for detecting MET amplification, they analyzed 232 inoperable gastric cancer patients who were treated with various firstline fluoropyrimidine-based regimens, which may have influenced treatment outcomes. A univariate analysis was conducted for OS and PFS, and only 170 patients were included in the PFS analysis without any information regarding MET amplification status and treatment regimens, which may cause biased results (26). Matsusaka et al. reported that MET gene amplification was associated with OS, but not with PFS in 150 patients who were uniformly treated with S-1 and cisplatin for metastatic or recurrent gastric cancer, which needs careful interpretation since patients received the same regimen and the MET amplification was assessed by real-time PCR (29). These studies applied an arbitrary threshold ≥5 copy number for identifying amplification without considering samples'tumor proportion. Having enough tumor tissues for the amplification assessment was important for accurate results by reducing the risk of normal cell dilution. With these limitations and results from RILOMET-1 trial, the prognostic impact of MET amplification has not been established, and our results suggest that MET amplification is not a prognostic predictor in patients with unresectable or recurrent gastric cancer who were treated with palliative FP.
MET inhibitor monotherapy, including tivantinib and foretinib, showed modest efficacy in unselected patients with metastatic gastric cancer (34-36). In a phase II study of tivantinib, there was no obvious relationship between drug efficacy and biomarkers, including MET gene amplification,and expression of c-MET, p-MET, and hepatocyte growth factor (34). AMG337 monotherapy showed remarkable response rate (5 of 10; 50%) in patients with METamplified gastroesophageal cancer in a phase I trial (37),however, the phase II study was terminated early due to efficacy and safety issues. The combination of chemotherapy and monoclonal antibodies blocking the c-MET pathway could not meet their primary endpoints in patients with MET-positive disease according to IHC(33,38,39). MET gene aberrations might not be the single driver of oncogene addiction, or other appropriate biomarkers might exist for MET inhibitor. Our results showed no prognostic value of MET gene amplification,which supports these hypotheses. However, given the clear association between clinical aggressiveness and MET amplification or protein overexpression in resectable gastric cancer, the MET pathway could have a pivotal role in the development of metastasis or recurrence from resectable diseases.
Our study may have some possible limitations because it is a retrospective single centre study, although part of our cohort was prospectively collected. Furthermore, selecting patients who had enough tumor tissues to assess and received uniform chemotherapy may have led to potential bias, even though these processes were essential for accurate results. However, after excluding inappropriate patients for analysis, our study investigated the largest dataset assessing MET amplification in metastatic or locally advanced unresectable gastric cancer. For applying our results in cases with unsuitable tumor proportion, tumor microdissection is essential; we believe that this could make our results more reliable and useful in clinical applications.
Conclusions
We found that MET amplification was not a prognostic predictor in patients with unresectable or recurrent gastric cancer who were treated with palliative FP, indicating that aberrant MET signaling pathway might not be the main driver in locally advanced or metastatic gastric cancer.Further validation is warranted to determine the clinical significance of MET amplification in patients with gastric cancer in the palliative setting.
Acknowledgements
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health &Welfare, Republic of Korea (No. HI12C1785).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
杂志排行
Chinese Journal of Cancer Research的其它文章
- Chinese guidelines for diagnosis and treatment of melanoma 2018 (English version)
- An update on biomarkers of potential benefit with bevacizumab for breast cancer treatment: Do we make progress?
- Association of cancer prevention awareness with esophageal cancer screening participation rates: Results from a populationbased cancer screening program in rural China
- FAT1, a direct transcriptional target of E2F1, suppresses cell proliferation, migration and invasion in esophageal squamous cell carcinoma
- A 18FDG PET/CT-based volume parameter is a predictor of overall survival in patients with local advanced gastric cancer
- Radiomics-based predictive risk score: A scoring system for preoperatively predicting risk of lymph node metastasis in patients with resectable non-small cell lung cancer
