Medical Knowledge Extraction and Analysis from Electronic Medical Records Using Deep Learning
2019-07-12PeilinLiZhenmingYuanWenboTuKaiYuDongxinLu
Peilin Li, Zhenming Yuan*, Wenbo Tu, Kai Yu, Dongxin Lu
Engineering Research Center of Mobile Health Management,Ministry of Education, Hangzhou Normal University,Hangzhou 311121, China
Key words: medical knowledge extraction; electronic medical record; named entity recognition; medical relation extraction; deep learning; bidirectional long short-term memory; conditional random field
W ITH the application of Internet technology in the field of medicine, great amount of electronic medical records(EMR) with rich medical knowledge have emerged. The electronic medical records are generated by medical staff for the purpose of recording the process of developing medical treatment with patients, including presentation of the disease, laboratory and imaging findings, therapeutic regimens and diseases progress, and so on. It implies the potential relation between the disease characteristics, drug usage and outcome of treatment.1The analysis and mining of these implicit knowledge can provide constructive help for medical decision-making and also provide data foundation that subsequently support the establishment of medical knowledge graph and visual display.
Electronic medical records are usually full of unstructured text, which can be post-structured by natural language processing to obtain the required medical knowledge.2Named entity recognition (NER) and medical relation extraction are two basic steps of the medical knowledge extraction. The logical relation between these two steps is series connection.
The main purpose of NER is to identify entities with specific meaning from the text. The original NER task mainly adopted a rule-based approach, which complete entity recognition and extraction tasks by formulating entity dictionaries and setting grammar rules. However, rules and grammars require supports of domain experts. The labor costs are considerable,and the flexibility is poor. The set grammar rules do not necessarily adapt to the new types of named entities. Later, some researchers have defined a NER task as the tagging task of the word sequence3and applied the machine learning method to the NER task. Character sequence tagging specifies Chinese characters in a sentence as a label, such as the “BIESN” annotation method.4The typical machine learning methods to deal with sequence tagging tasks are hidden Markov model(HMM) and conditional random field (CRF). Ye F5used CRF and combined with multiple feature templates for NER tasks of electronic medical records. Li W6combined CRF with rule method, which first completed sequence annotation with CRF, and then optimized the results of CRF with rules based on decision tree and clinical knowledge so as to achieve entity recognition of electronic medical records. However, CRF method is strongly dependent on manually extracted grammatical features. As the result, the setting and selection of feature templates by researchers will directly affect the accuracy of NER.
Since the concept of relational extraction was initially introduced at the MUC-7 conference in 1998, it has been widely studied and developed in a variety of fields,such as finance, education, medical care and so on.7-9In the field of medicine, medical relation extraction (MRE)is used to extract medical relations among items of entities from electronic medical record. The extraction of medical relations in the single sentence of electronic medical records was the preliminary step in a study.10Due to the lack of classification criteria for relation types, six entity relation types are defined in advance,and manually selected features are used to extract medical relations through support vector machine (SVM)classifier. In the task published in 2010, I2B2 dataset proposed eight different relation type schemes,11,12which established a scientific classification standard for the field of medical entity relation extraction. Considering the similarity of text types between contexts surrounding medical entities, Rink B13used SVM classifier for classification, and the best F1 classification results at that time were obtained in I2B2 2010 dataset. However,the context information related to entities is diverse,which leads to the sparsity of feature vector representation. In order to solve this problem, the entity semantic relation in UMLS is used as the classification feature to improve the performance of medical entity relation recognition task.14In addition, Lv X15proposed a deep learning method for feature engineering, which uses automatic encoder to solve the problem of data sparsity and uses CRF as the classification model to classify the relation between entity pairs.
In recent years, deep neural network (DNN) has been widely used in various fields. The GloVe word embedding model16brings great convenience to the NLP task based on DNN. Meanwhile, the recurrent neural network (RNN) with timing characteristics has been increasingly used in the field of NLP, and its effect has been proved generally better than other methods.17Therefore, based on the recent deep learning algorithm,we built two application scenes of BiLSTM-CRF model in NER and MRE tasks, and validated it on the I2B2 2010 public dataset, where it showed good results.
MATERIALS AND METHODS
Long short-term memory
As a time-series recurrent neural network, long shortterm memory (LSTM) is suitable for processing and predicting important events with relatively long intervals and delays in the time series. Additionally, it can solve the gradient explosion problem when recurrent neural networks (RNNs) are used for long-term sequence predictions.
There are three control gates in each neuron of LSTM, namely, the input gate, forgetting gate and output gate. The output of the previous unit enters the LSTM unit and it is judged whether it is useful according to the cell. Only useful information is left, and the nonconforming information is forgotten by the forgetting gate. Equations (1) through (5) represent the parameter update process, where σ represents the sigmoid function, ht-1represents the output of the previous unit of the LSTM, and htrepresents the current output. The structure of the LSTM unit is shown in Figure 1.

BiLSTM uses two LSTM networks to propagate the sequence forward and backward, respectively, so that the context information of the sequence can be extracted more comprehensively.
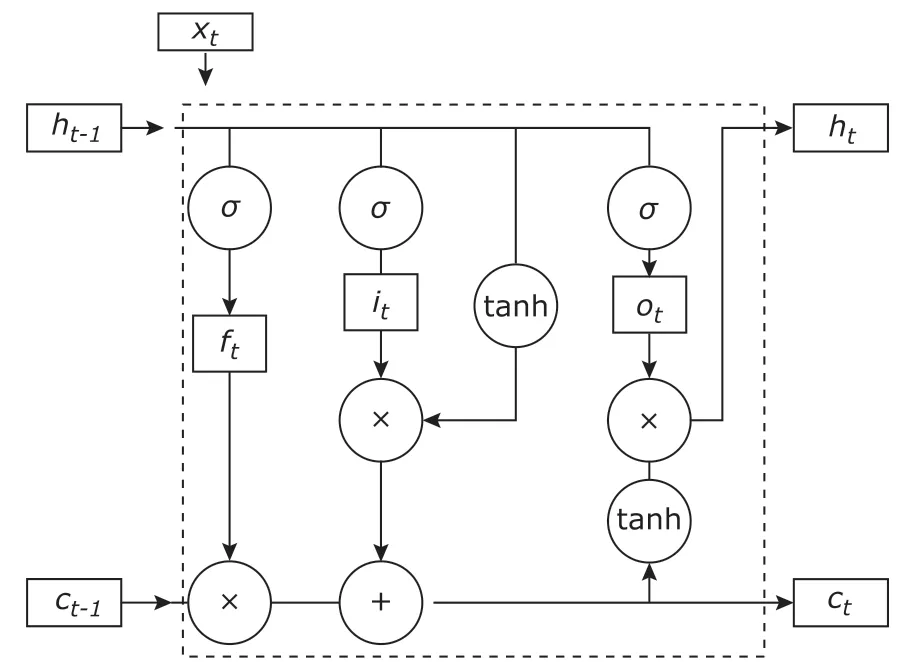
Figure 1. LSTM neuron structure. The tanh is a classical neural network nonlinear activation function. The input gate, the forgetting gate, and the output gate in the LSTM unit are defined as i, f, and o, respectively. Ct is the state of the storage unit at the current time, and equation (4)represents the process of the state transition of the memory unit. The current state is calculated by the previous time state Ct-1, the result of the forgotten gate ft, and the input gate it of the current time LSTM unit.
Data preprocessing
The data used in this study is the unstructured English EMR text in the I2B2 2010 evaluation task12with the officially labeled medical entity category and medical entity relation category. The dataset divides medical entities into three categories: medical problems, treatment, and tests. It classifies the relations contained between medical entities into eight categories, as shown in Table 1.
This study took a sentence in the EMR as a sequence and tried to identify and classifies all entities and medical relations contained in the sentence. As shown in the example in Figure 2, the sentence contains three entities, tests, medical problems and treatment, in which “CNIS” and “Carotid stenosis” constitute the medical relation of “TeRP”.
In the input section, we used Stanford’s GloVe word embedding model to convert EMR text data into 100-dimensional word vector data. We used the 822MB corpus from Wikipedia to combine the English electronic medical record corpus of I2B2 2010 as thetraining corpus of the GloVe word vector.

Table 1. Categories of medical relations and descriptions

Figure 2. Text and labeling example of electronic medical record (EMR). The example has three parts. The first part is the sentence containing the entity in the medical record, where“CNIS” is the abbreviation of calcineurin inhibitors (CnIs)and steroids treatment for vascular disease. The middle part shows the line:column number of the entities in the record and its category. The last part is the relation, TeRP, between the entities (CNIS and carotid stenosis) in the sentence.
BiLSTM-CRF in NER
In the NER task, we classified the entities by the strategy of sequence labeling. We used a sentence as a sequence of model input and used the joint labeling strategy of “BIESO” and entity categories for the entities in the sentence. The model first used GloVe to transform the word into word vector. Then it got the hidden layer state of the node through BiLSTM, and finally carried out the joint probability distribution through the CRF layer. The model structure is shown in Figure 3.
BiLSTM-CRF in MRE
The result of NER is one of the input parts of the MRE task. So, the MRE task is in tandem with the NER task.The smallest input unit of the MRE model is the entity.Each medical entity is a set of word vectors containing the entity itself and its context information. In order to understand the model more clearly, Figure 4 shows the neural network structure of a single entity(carotid stenosis) in the model. For each medical entity included in the sentence, we set a context window for it,which is demonstrated using a window size with a fixed context of 2 words.
The word vector sequence is the input of the BiLSTM layer for learning. Considering that the length of the entity is not fixed, we input the start and stop position information of the entity after the BiLSTM layer, and only extract the hidden layer state of the corresponding entity position in the previous layer. The purpose of this step is to avoid the similarity of the feature information of adjacent entities and to affect the training effect of the model.
Figure 5 shows the complete BiLSTM-CRF medical relation extraction model. After passing through the neural network process shown in Figure 4, each entity performs probability calculation through the CRF layer to obtain a corresponding medical relation classification. The purpose of this layer is to transform the classification problem of a single entity into a sequence classification problem, and to link the feature combinations between entities. In fact, the model can be seen as a CRF model optimized for feature engineering by word vector and BiLSTM.
RESULTS
As mentioned earlier, this study uses the English electronic medical record data published in the I2B2 2010 evaluation meeting to conduct experiments. Of the 871 marked medical records, 397 were used for training and 477 were used for testing.18The purpose of the experiment was to identify the correct physical and medical relations from the unstructured electronic medical record text. This study uses precision (P),recall (R) and F1-measure as indicators of evaluation.F1-measure is a weighted harmonic averaging of the precision and the recall. It is a commonly used evaluation standard in the field of Information Retrieval and is often used to evaluate the quality of a classification model. Let the result set of the model output be y, and the result set of the manual label be y. The formula for the evaluation criteria is as follows:

Figure 3. BiLSTM-CRF structure of named entity recognition(NER). “l1”, “r1”, “e1”, etc., represents the different network layers in the model. The CRF layer is a combination label, where “B”, “I”, “E”, “S”, “O” are word segmentation labels and “Te”,“P”, “Tr” are entity category labels. BiLSTM, bidirectional long short-term memory; CRF, conditional random field.

Figure 4. Single entity neural network structure of medical relation extraction(MRE). “TeRP” in the CRF layer is a relation category label.
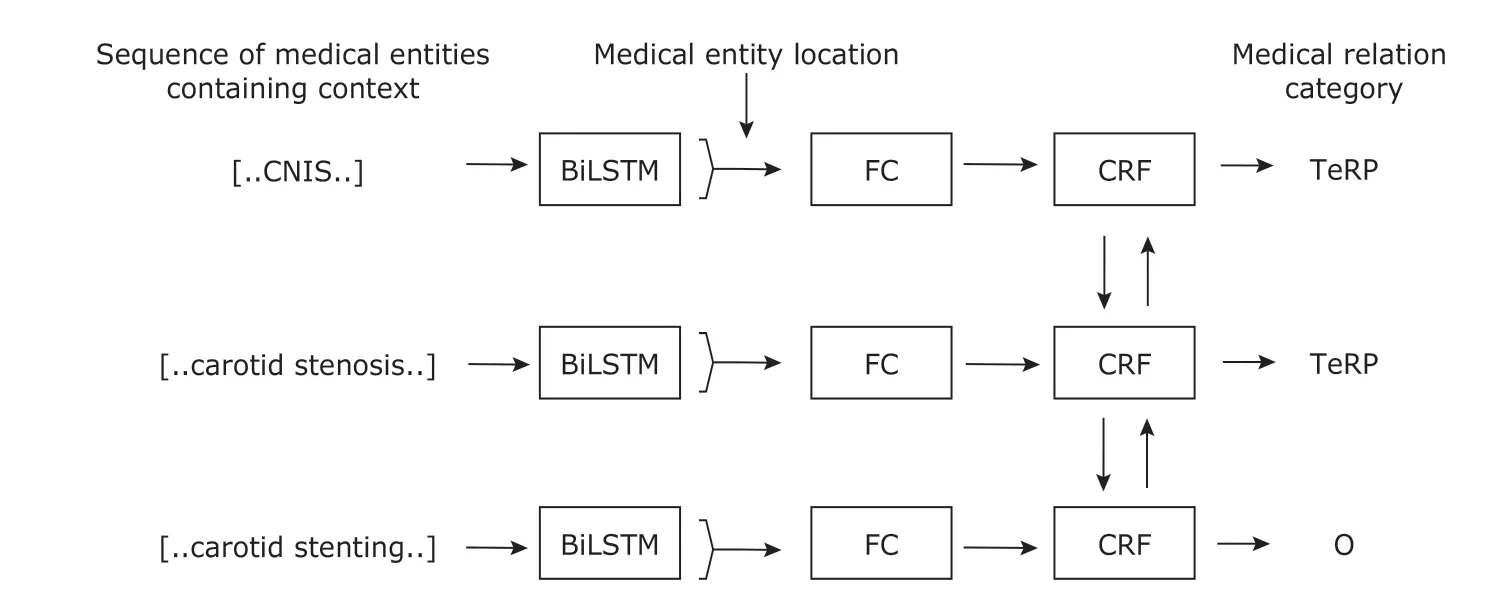
Figure 5. BiLSTM-CRF structure of MRE. FC is the fully connected layer.

The result of NER
The experimental results of NER are shown in Table 2. We compared the model in this study with other well-performing models on the dataset. The SVM and CRF model were proposed by Jiang M,19which useed the Systematized Nomenclature of Medicine (SNOMED)as an auxiliary resource. The LSTM model was chosen for a more intuitive comparison. The results show that the BiLSTM-CRF model has better expressiveness in the dataset. Moreover, the model converged faster and avoided problems such as overfitting.
The result of MRE
The experimental results of MRE are shown in Table 3. We compared the BiLSTM-CRF model with other well-performing models. Among them, SVM13and maximum entropy (ME)20are both obtained by semi-supervised training. It is clear that the performance of our model is still better than others. From the implementation details, the classification of our model can effectively avoid the misclassification of non-entity texts. There will also be good performance in real scenes outside the dataset.

Table 2. The F1-measure of models in NER
DISCUSSION
From the experimental results, the BiLSTM-CRF model constructed in this study has made a significant improvement over the previous research. The F1-measure of NER is close to 0.88, and of MRE is close to 0.78. However, there still remain some problems. The model has categories with a low recognition rate in both tasks. Especially in MRE, there are three categories where the F1-measures were less than 0.4. The general reason for this problem is that these categories are too small in the dataset and was difficult for the model to understand their semantics. For example,in the sentence “He was started on p.o. steroids and to CMED for management of COPD exacerbation but he appeared in more respiratory distress overnight”,the “management” and “COPD exacerbation” should constitute a “TrWP” relation, but the model identified it as a “TrAP”. The reason lied in that the patient’s deterioration appears in the next short sentence, and the model didn’t perfectly learn the feature.
In addition, the effects of the model on several categories were higher than 0.9, of which “PIP” was all correctly classified. This is due to the large number of its examples and less interference.
This study constructed different structure of BiLSTM-CRF model and verified the good performance of deep learning model in medical knowledge extraction.This shows that the same model can be applied to different scenarios by reasonable modeling of the characteristics of the task. In view of the fact that clinical electronic medical records contain more domain experience knowledge, how to make corresponding adjustments and changes from the general method, and how to construct a more specific model will be questions that NLP needs to think about in the research field of clinical EMRs in the future. Besides, as two subtasks of MKE, the clinical accuracy of MRE is seriously affected by the results of NER. Although NER has a F1-measure close to 0.9, the error will be magnified during transmission. Therefore, when it comes to deeper research,it is imperative to further improve the effect of basic tasks in MKE.

Table 3. The F1-measure of models in MRE
Conflict of interest statement
The authors have no conflict of interests to disclose.
杂志排行
Chinese Medical Sciences Journal的其它文章
- Integrative Analysis Confirmed the Association between Osteoprotegerin and Osteoporosis
- A Survey of Surgical Patient’s Perception about Anesthesiologist in a Large Scale Comprehensive Hospital in China
- A Survey on Intelligent Screening for Diabetic Retinopathy
- Multi-Atlas Based Methods in Brain MR Image Segmentation
- Application of Mixed Reality Technology in Visualization of Medical Operations
- Constructing Large Scale Cohort for Clinical Study on Heart Failure with Electronic Health Record in Regional Healthcare Platform: Challenges and Strategies in Data Reuse
